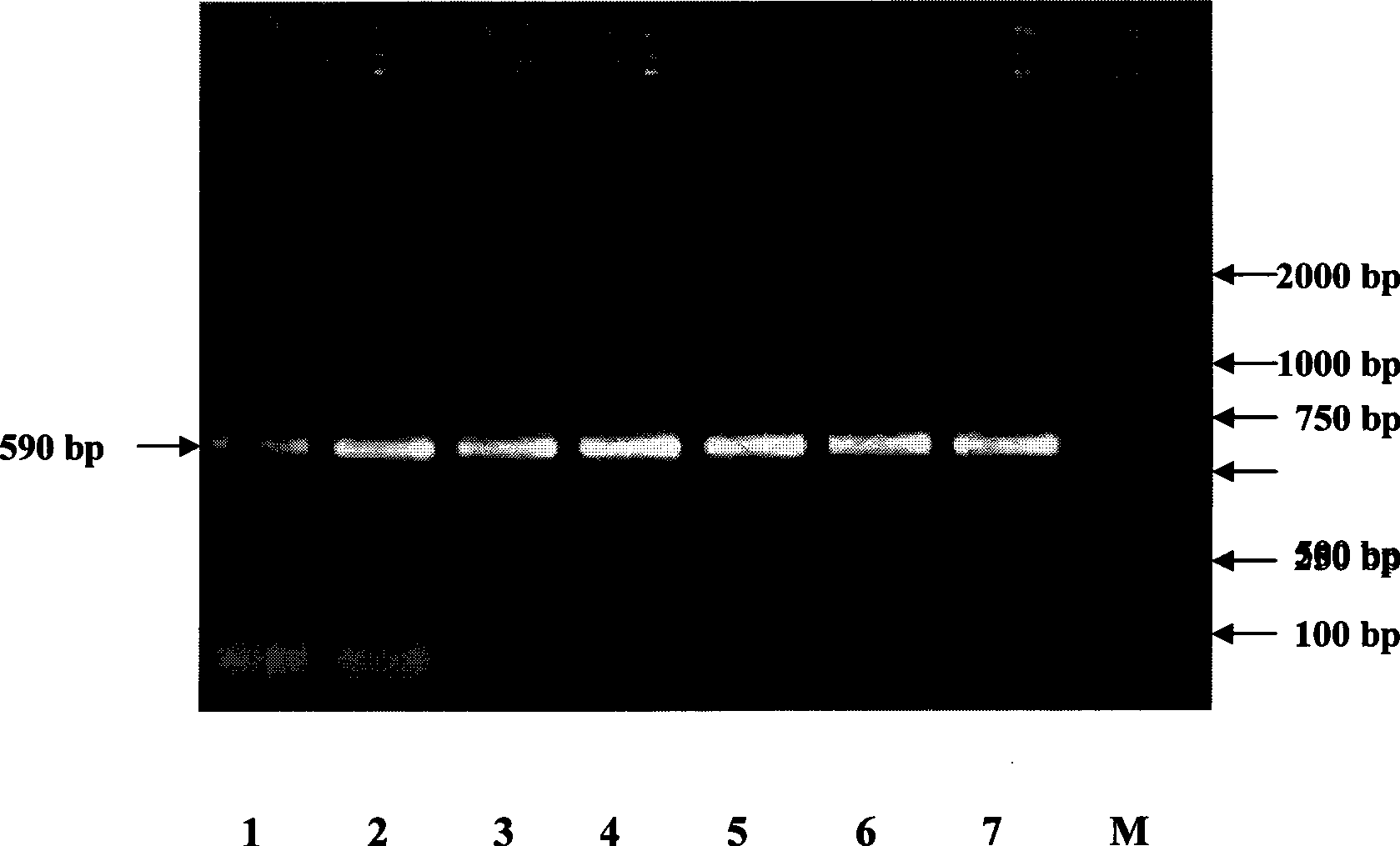Molecular marker method for identifying pepper anti-epidemic disease character
A technology for molecular markers and anti-epidemic diseases, applied in biochemical equipment and methods, microbial measurement/inspection, etc., to achieve simple and fast steps, improved selection efficiency, and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] 1. Artificially designed and synthesized nucleotide sequences as primers:
[0040] Forward primer: 5’-AGGTGGTCTTCAATGATCAGAGA-3’
[0041] Reverse primer: 5’-CCTGAGGCAAATTCCAAATCTC-3’
[0042] 2. Extraction of genomic DNA of pepper: A modified SDS method was used to extract genomic DNA of 7 pepper inbred lines A5, A11, B17, B23, B2, B32 and B19 (Gong Zhenhui, etc., using PCR to identify the trace DNA extraction method of transgenic plants [ J]. Journal of Northwest Agricultural University, 1997, 25(1): 45-47).
[0043] 3. Using the nucleotide molecules described in step 1 as primers, PCR amplification was performed on the genomic DNA of the 7 pepper inbred lines mentioned above. All 7 pepper inbred lines could amplify a fragment of 590bp in length ( figure 1 Agarose gel electrophoresis analysis of PCR amplification products of 7 pepper inbred lines. M: DNA Marker; 1: A5; 2: A11; 3: B17; 4: B23; 5: B2; 6: B32; 7: B19). The PCR amplification system is as follows:
[0044] (1)...
Embodiment 2
[0051] 1. Artificially designed and synthesized nucleotide sequences as primers:
[0052] Forward primer: 5’-AGGTGGTCTTCAATGATCAGAGA-3’
[0053] Reverse primer: 5’-CCTGAGGCAAATTCCAAATCTC-3’
[0054] 2. Extraction of pepper genomic DNA: The genomic DNA of 7 pepper inbred lines B3, P11, B4, P30, B12, P97 and P45 were extracted using modified SDS method.
[0055] 3. Using the nucleotide molecules described in step 1 as primers, PCR amplification was performed on the genomic DNA of the 7 pepper inbred lines mentioned above. All 7 pepper inbred lines could amplify a fragment of 590bp in length ( image 3 Agarose gel electrophoresis analysis of PCR amplification products of 7 pepper inbred lines. M: DNA Marker; 1: B3; 2: P11; 3: B4; 4: P30; 5: B12; 6: P97; 7: P45). The PCR amplification system is as follows:
[0056] (1) The volume of the reaction solution is 25μL, and the composition and content of the articles used are:
[0057] 10 times PCR buffer 2.5μL, 25mmol·L -1 MgCl 2 2.0μL, 5U·...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com