Epithelial type cadherin expressed gene CDH1 mutation detection kit and application thereof
A detection kit, kit technology, applied in the field of genetic diagnosis and consequence assessment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
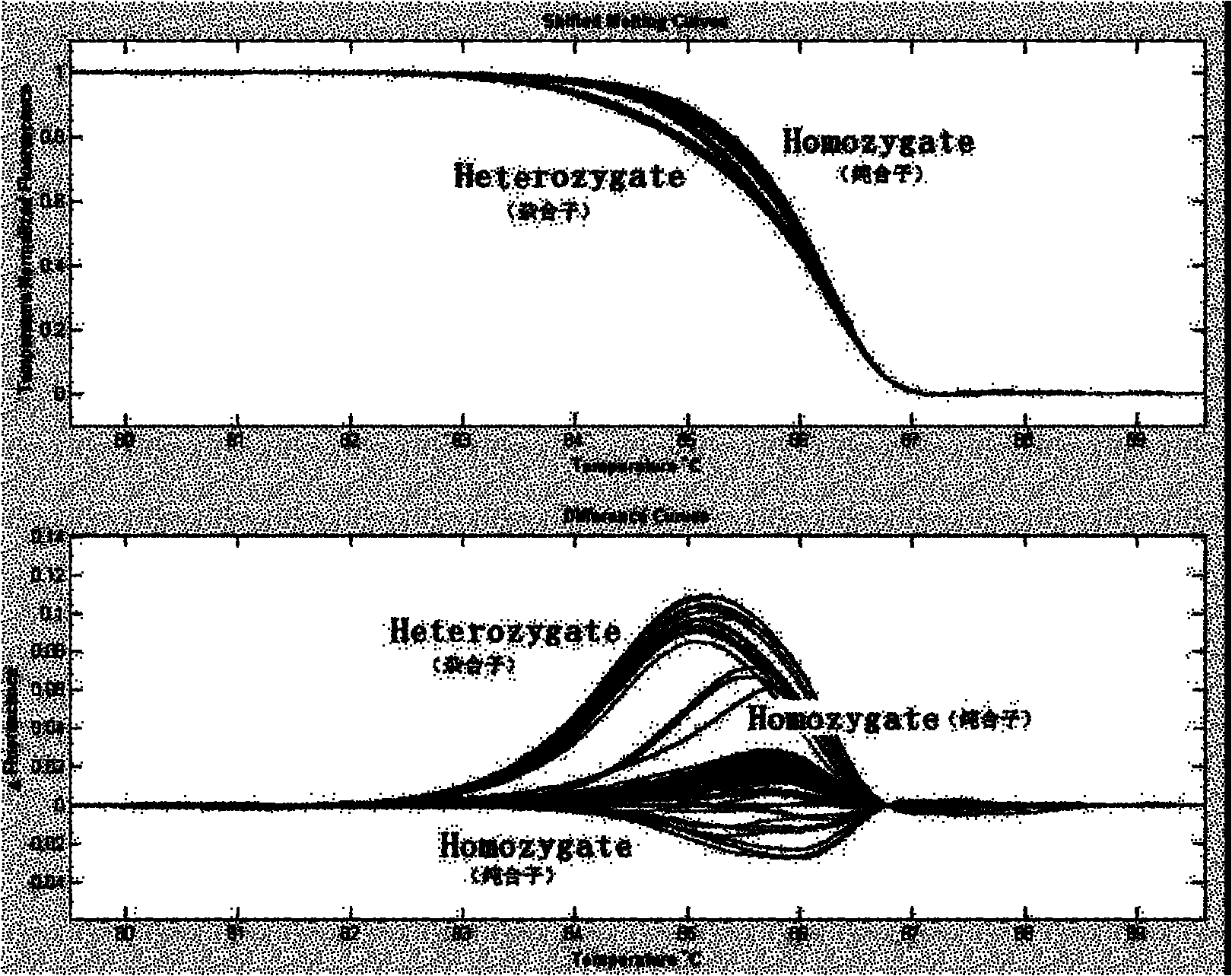
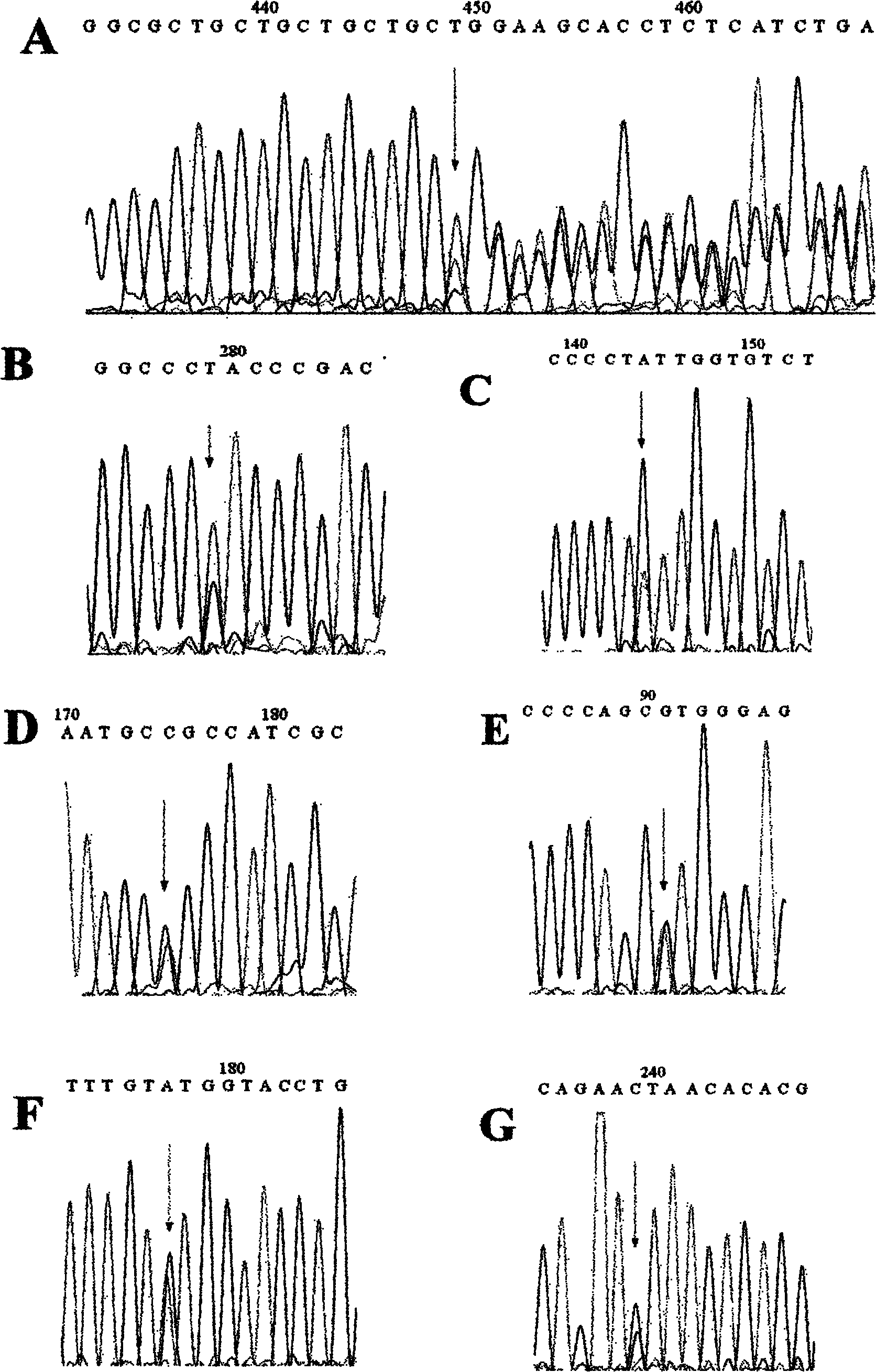
[0032] Example 1: Systematic screening and case-control analysis of CDH1 gene mutation in Chinese patients with gastric cancer
[0033] A total of 238 patients with gastric cancer in Jiangsu, Anhui and Zhejiang regions of China and 240 normal controls were selected for peripheral blood extraction and DNA extraction. For exon 2-12 and 14-16 of CDH1 gene, primers were designed according to Table 1 for PCR amplification and high-resolution melting curve (HRM) analysis. The reaction system is: Genomic DNA (100ng / μl) 1.0μl, dNTP Mixture (2.5mM each) 0.8μl, 10×PCR buffer (Mg 2+ free) 1.0μl, MgCl 2 (25mM) 1.0μl, upstream primer (10μM) 0.2μl, downstream primer (10μM) 0.2μl, dimethyl sulfoxide (DMSO) 0.4μl, ddH 2 O 4.32 μl, LC Green dye 1.0 μl, Taq DNA polymerase (5 Unit / μl) 0.08 μl. Reaction conditions: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 sec, renaturation at 60°C (corresponding annealing temperature) for 30 sec, extension at 72°C for 40 sec, 40 cycles; ...
Embodiment 2
[0039] Example 2: Analysis of the impact of detected CDH1 gene missense mutations on protein function
[0040] Using SIFT analysis (the Sorting Intolerant from Tolerant, http: / / sift.jcvi.org / ) and PolyPhen (=Polymorphism Phenotyping, http: / / genetics.bwh.harvard.edu / pph / ), analysis of missense mutations caused Effects of single amino acid changes on CDH1 protein function. The score threshold given by SIFT is 0.5, which means that those 0.5 are tolerable. The PolyPhen tool calculates the difference between the PSIC score of the encoded amino acid before and after the mutation to determine the impact of the mutation on the structure and function of the protein. When the difference exceeds 1.5, it is considered a mutation that changes the function. SIFT analysis in this study showed that the detected CDH1 c.1888C>G(L630V) mutation scored 0.02, while the PolyPhen score difference reached 1.748, so this mutation may affect the function of CDH1 protein. But V202I and T340A are only...
Embodiment 3
[0043] Example 3: Functional Consequence Analysis of Detected CDH1 Gene Promoter Region Mutations
[0044] The functional consequences of mutations in gene promoter regions were detected using a dual-luciferase reporter gene assay. Various mutant plasmids in the promoter region of PGL3-CDH1 were constructed, transfected into Hela cells, and the promoter activity intensity of each mutant gene was evaluated according to the relative luciferase activity of the expressed products.
[0045] (1) Construction of various PGL3-CDH1 promoter region mutant plasmids:
[0046] According to the sequence of CDH1 gene promoter region, design primers with double restriction sites. PCR amplification included a region from -345 to +271bp from the transcription initiation site. The upstream primer is PF: 5'-ATGC CTCGAG CCATTCTCCAAAACGAACAAAC-3' (SEQ ID NO: 39), 'ATGC' is a protection sequence,' CTCGAG ' is the recognition sequence of XhoI endonuclease. The downstream primer is PR: 5'-ATGC ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com