Method for detecting oral squamous-cell carcinoma and method for suppressing the same
A squamous cell carcinoma and detection method technology, applied in the field of cancer detection, can solve the problem that the prognosis has not been improved, and achieve the effect of inhibiting proliferation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0044] Dry spots can be prepared, for example, by dripping endless BACDNA or the like onto a substrate using a spotter, forming a plurality of spots, and then drying the spots. As the spotter, an inkjet printer, dot matrix printer, or dual jet (registered trademark) printer can be used, and an inkjet printer is preferably used. For example, GENESHOT (manufactured by Gaishi Co., Ltd., Nagoya City, Japan) and the like can be used.
[0045] In this way, a desired DNA-immobilization substrate can be prepared by immobilizing infinite BAC DNA or the like on a substrate, particularly preferably a solid substrate.
[0046] In addition, as one of means for directly detecting the deletion of the PRTFDC 1 gene, Southern blotting can be cited. The Southern blotting method is a method of separating and fixing the genomic DNA obtained from the sample, and detecting the presence of the gene in the sample by detecting the hybridization between the genomic DNA and the PRTFDC1 gene.
[0047] ...
Embodiment 1
[0100] Example 1: Alterations in genes in oral squamous cell carcinoma
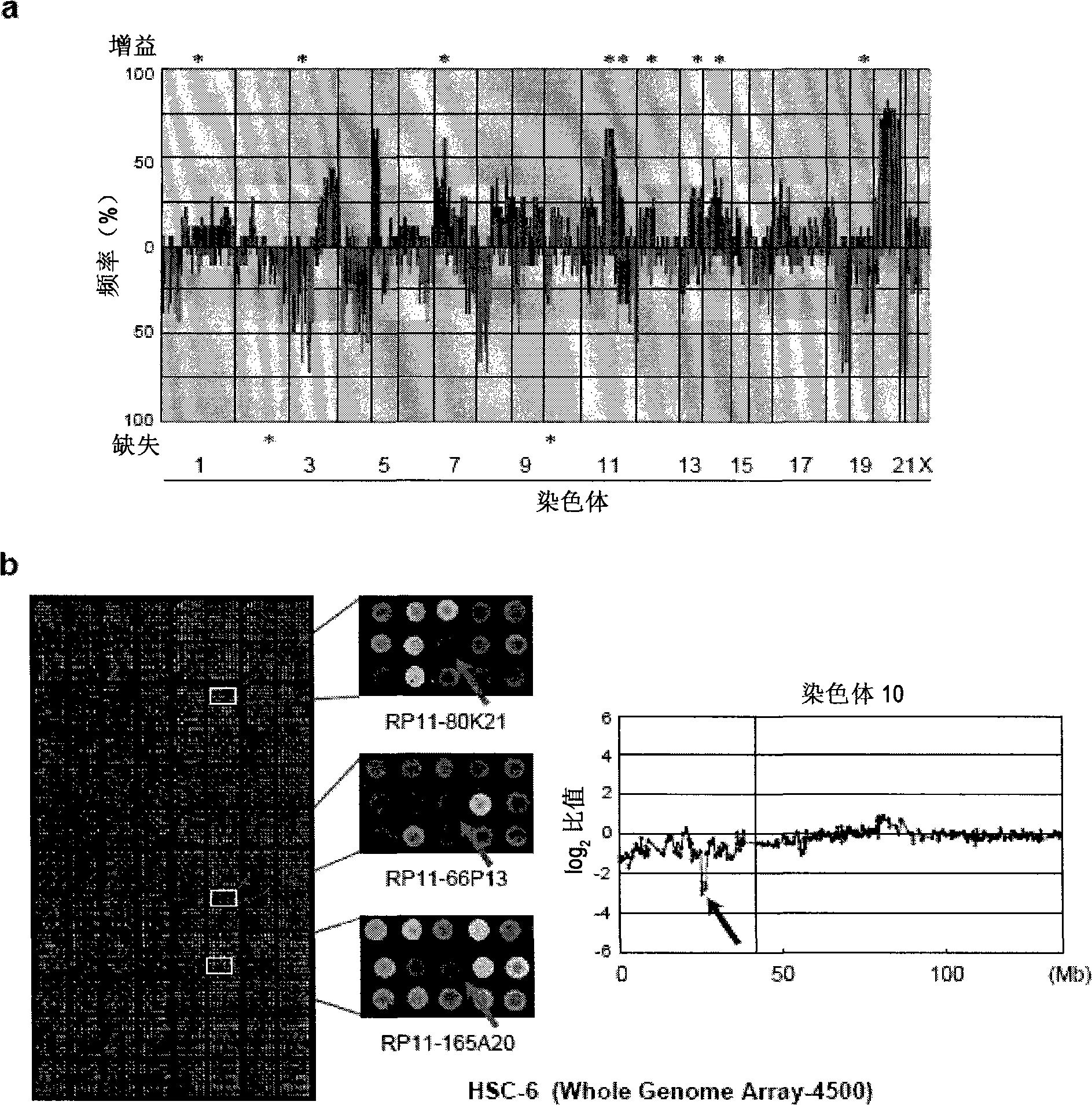
[0101] In order to detect novel gene changes in OSCC, 18 OSCC cell lines (OM-1, OM-2, TSU, ZA, NA, Ca9-22, HOC-313, HOC- 815, HSC-2, HSC-3, HSC-4, HSC-5, HSC-6, HSC-7, KON, SKN-3, HO-1-N-1, KOSC-2) prepared genomic DNA, using MCG CancerArray-800 and MGC Whole Genome Array-4500 for CGH array analysis ( figure 1 a). The genomic DNA extracted from a normal oral epithelial cell line (RT7) was used as a control to be labeled with Cy5; the genomic DNA prepared from an oral squamous cell carcinoma cell line was used as a test DNA to be labeled with Cy3. Specifically, DpnII-digested genomic DNA (0.5 μg), respectively, in 0.6 mM dATP, 0.6 mM dTTP, 0.6 mM dGTP, 0.3 mM dCTP, 0.3 mM Cy3-dCTP (oral squamous cell carcinoma cells) or 0.3 mM Cy5 - In the presence of dCTP (normal cells), the cells were labeled with the BioPrime ArrayCGH Genomic Labeling System (manufactured by Invitrogen). Cy3 and Cy5 labeled dCTP wer...
Embodiment 2
[0114] Example 2: Isolation of genes contained in the 10p12 chromosomal deletion region in oral squamous cell carcinoma
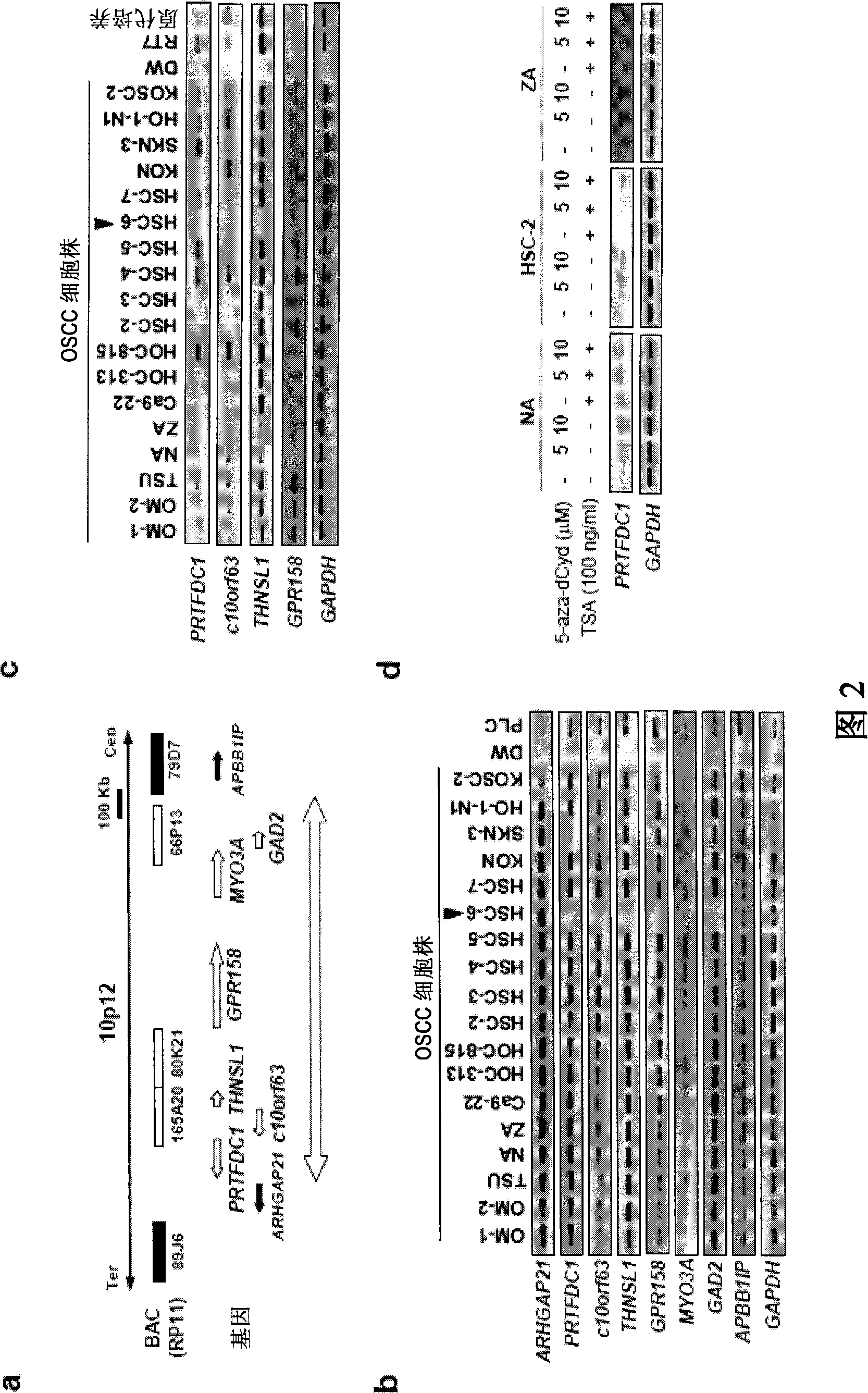
[0115]To determine the genes involved in the homozygous deletion region of chromosome 10p12 in oral squamous cell carcinoma (HSC-6), first, using genomic PCR from HSC-6 cells, the extent of the homozygous deletion region was determined. The primer sequences used in genomic PCR are shown in Table 3.
[0116] table 3
[0117] Supplementary Table 2. Primer sequences used in this study
[0118]
[0119] From the results of the CGH array and the human genome database (http: / / genome.ucsc.edu / ), it can be confirmed that the deletion region is a deletion of up to 2.95Mb ( figure 1 b. Figure 2a). Then, it was possible to confirm the presence of 7 genes in this region.
[0120] Among them, only in HSC-6 cells (1 / 18, 5.6%; Fig. 2b), can confirm the homozygous deletion of PRTFDC1 gene, c10 or f63 gene, THNSL1 gene, GPR158 gene, MYO3A gene, GAD2 gene, and ARHGAP2...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com