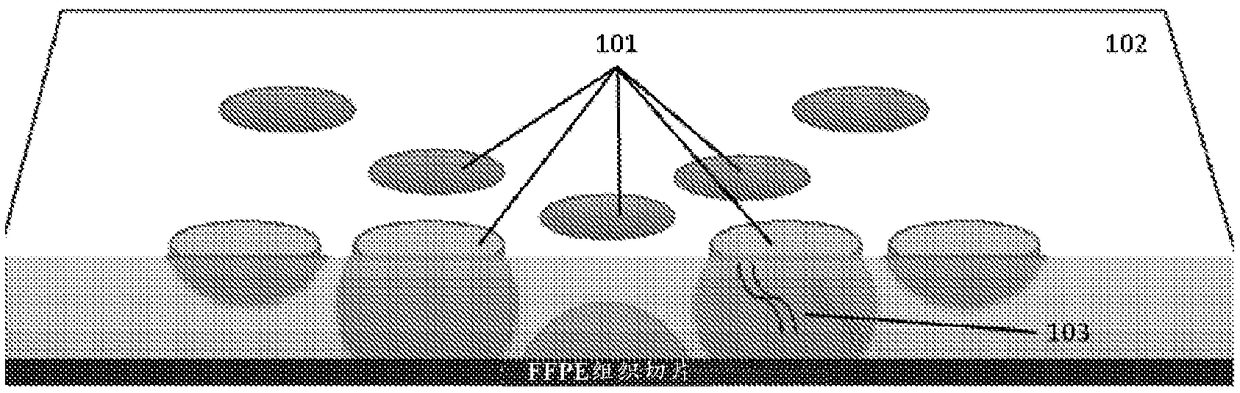
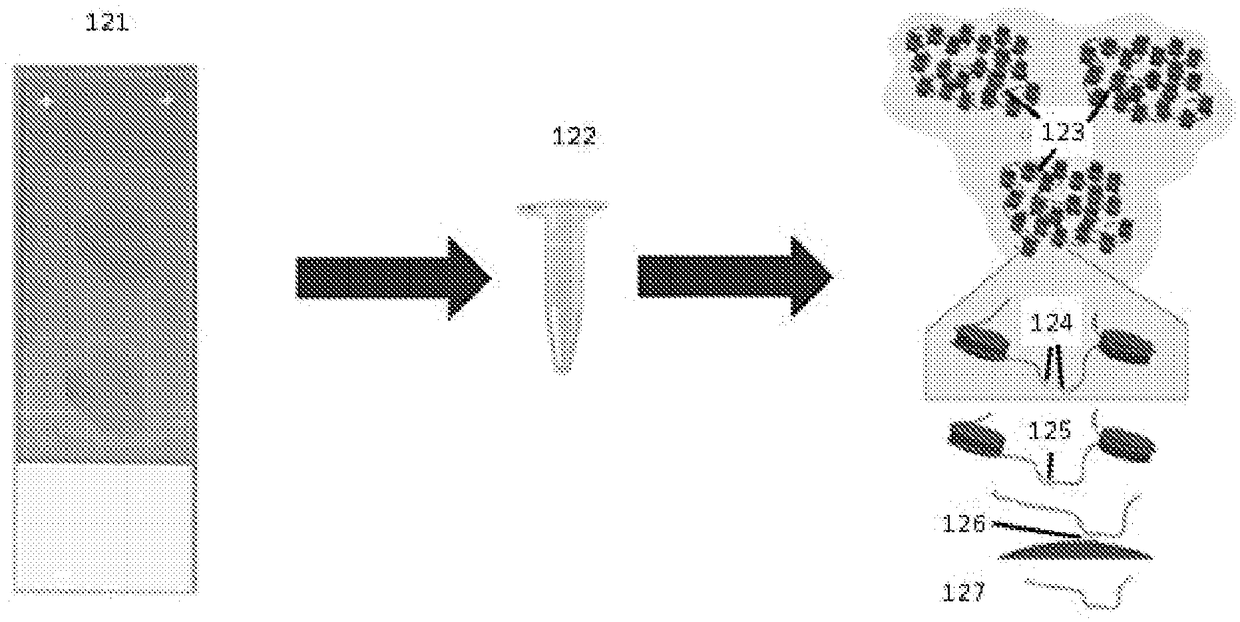
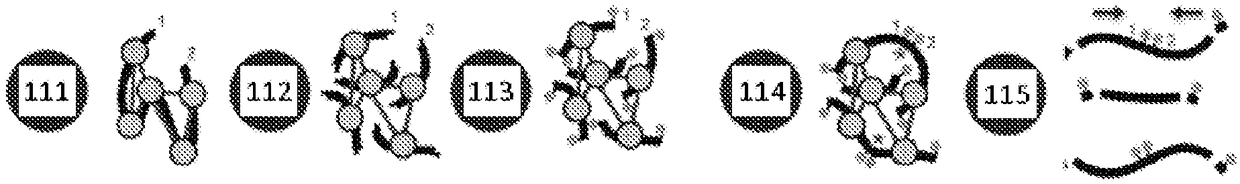Recovering long-range linkage information from preserved samples
A sample and nucleic acid technology, applied in the field of recovering long-range linkage information from preserved samples, which can solve problems such as DNA damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0274] Example 1. Generation of read pair libraries from FFPE samples
[0275] AJ GIAB (‘Genomes in a Bottle’) samples GM24149 (father) and GM24385 (son) were procured from HorizonDiscovery. Cell lines have been previously embedded in FFPE. Sections approximately 15-20 μm thick, each containing approximately 3x10 5 cells. Sections were washed with xylene to remove paraffin. Xylene was removed by washing the sections with ethanol. Released tissue samples were subsequently resuspended in detergent buffer. Samples containing the nucleic acid are then subjected to end ligation, which involves digestion of the DNA with a restriction enzyme (MboI in this example), followed by filling in the resulting overhangs with biotinylated nucleotides. Join the flat ends together, then release the joined ends. Biotinylated fragments are obtained and end-sequenced, and pairs of reads are obtained to indicate that each mapped contig is physically linked to a common nucleic acid molecule i...
Embodiment 2
[0281] Example 2. Phase determination from FFPE-Chicago library
[0282] The sequencing data generated in Example 1 was used to determine phasing information for sets of SNPs known to be in the starting GIAB samples. In other words, sequencing data is used to determine whether multiple sets of SNPs are present on the same or different DNA molecules. These data were then compared to known sequences of GIAB samples to determine the accuracy of the phase calls.
[0283] Each bin in Table 3 shows the number of SNPs found, and they are consistent until the size of the next bin. For example, the first row shows that between 0-10,000, 132,796 SNPs were found, and 99.059% were in the correct phase. High consistency (>95%) can be seen until about 1.5MB (except for 70-80kb bins (which miss 1 of 13) and 1.1-1.3MB bins (which miss 2 of 15 indivual)). Seven of the seven SNP pair phases were correctly called within the 1.7-1.9 MB range.
[0284] From these data it can be concluded th...
Embodiment 3
[0288] Example 3. Improved DNA extraction
[0289] Changing the detergent buffer from SDS-containing buffer to triton X-containing buffer and visualization of the precipitate described in Example 1 resulted in increased DNA extraction. Subsequent library analysis revealed that this library had increased complexity compared to the libraries described in Examples 1 and 2, while maintaining a high level of long reads. The results are shown in Table 4.
[0290] Human Sample 1 data were collected from GIAB samples processed as described in Example 1 (blunt end ligation on FFPE samples). All DNA from this sample was used for library preparation.
[0291] Human sample 2 data was collected from a second GIAB sample processed as described in Example 1 (blunt-end ligation on FFPE samples). All DNA from this sample was used for library preparation.
[0292] Human sample 3 data was collected from a third GIAB sample processed as described in Example 1 (blunt-end ligation on FFPE sam...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
| surface area | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com