Method for predicting cancer synthesis lethal gene pairs based on decision tree and linear regression model
A linear regression model and synthetic lethal technology, applied in genomics, instruments, biological systems, etc., can solve problems such as high cost, missing synthetic lethal gene pairs, and difficulty in synthesizing lethal gene pairs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0032] The technical solutions of the present invention will be described in detail below in conjunction with the accompanying drawings.
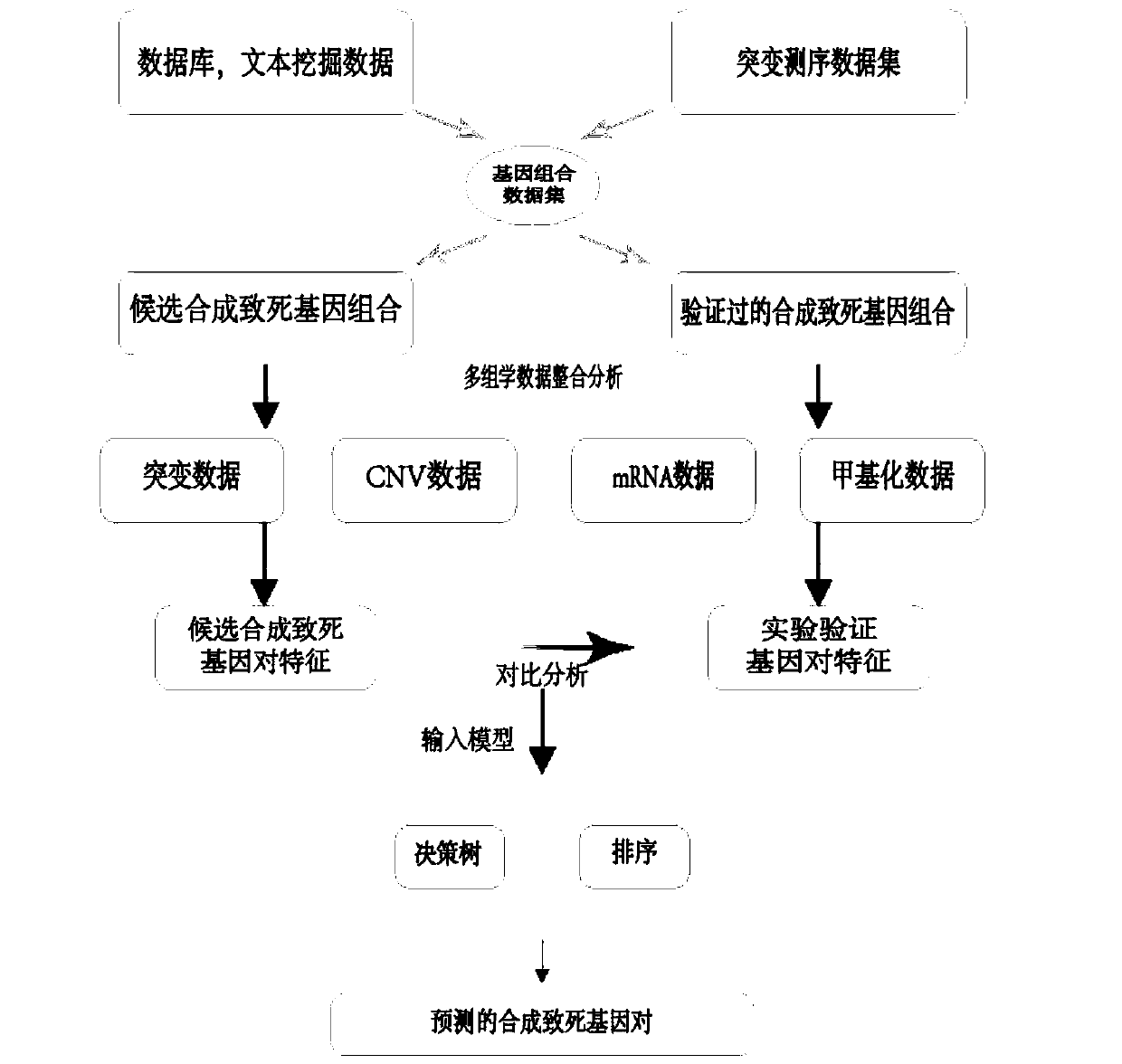
[0033] The method for predicting the synthetic lethal gene pair of cancer based on decision tree and linear regression model, specifically comprises the following steps:
[0034] 1) Extract multi-omics data from high-throughput sequencing data, and preprocess it into a matrix format, including gene name, sample name and corresponding quantitative data; the multi-omics data includes gene mutation, mRNA expression, DNA The medical big data of methylation and copy number variation can be analyzed from the perspective of a single cancer type or pan-cancer. Each feature of each cancer type is integrated and processed into a data matrix. The row name is gene name, and the column name is sample Numbering.
[0035] 2) Effective data screening is performed based on the multi-omics data in 1), specifically integrating the original data and removing ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com