An integrated method and system for identifying functional patient-specific somatic aberations using multi-omic cancer profiles
A specific and patient-specific technology, applied in genomics, biological systems, proteomics, etc., can solve the problems of ignoring the functional heterogeneity of interaction links, ignoring limitations, and translating to other tissue and pathological backgrounds
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
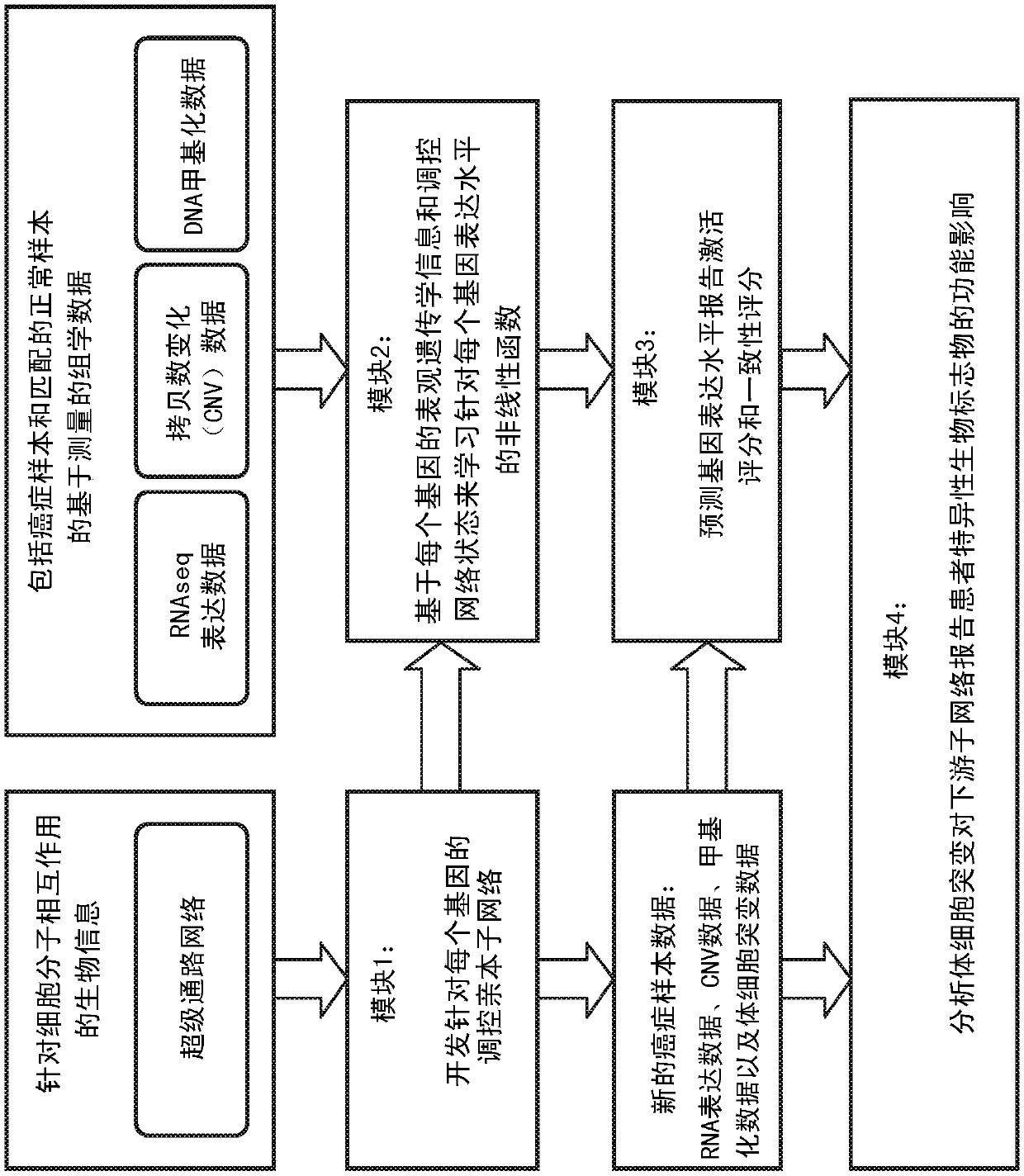
[0052] The present invention provides a method for integrating multi-omic bioinformatics and various sources of molecular measurement data into a unified web-based computational method for providing patient-specific gene expression predictions and identifying significant deviations in gene expression levels from expected levels and inconsistent systems and methods. Refer below Figure 1-12 The present invention is described in further detail.
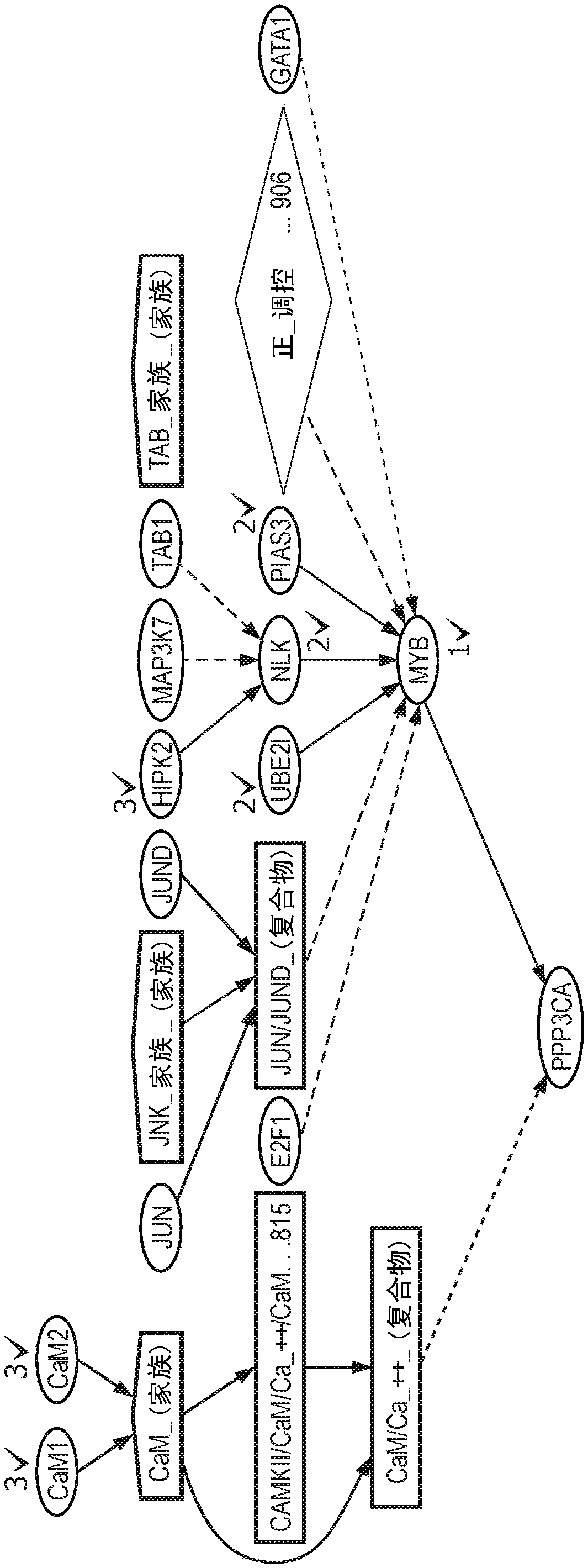
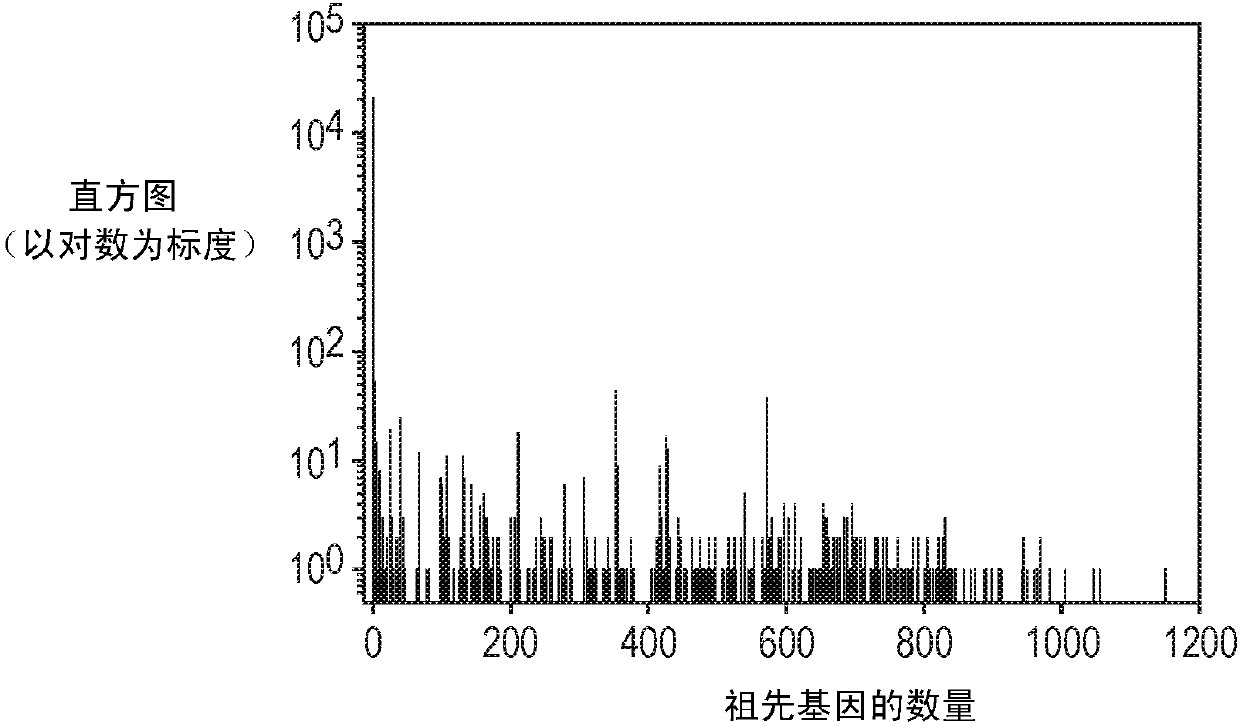
[0053] According to an embodiment of the present invention, by figure 1 The steps or modules outlined in to illustrate a flowchart presenting an overall block diagram of the method for providing patient-specific gene expression predictions, identifying significant deviations and inconsistencies in gene expression levels from expected levels, and reporting patient-specific biomarkers thing. Such as figure 1 As shown, the method consists of four major sequential steps or modules to identify and report potential somatic aberrations dri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com