System and method of analyzing association change pattern from multi-omics data
A technology of omics data and patterns, applied in the analysis of 2D or 3D molecular structure, informatics, bioinformatics, etc. The effect of applicability and strong compatibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
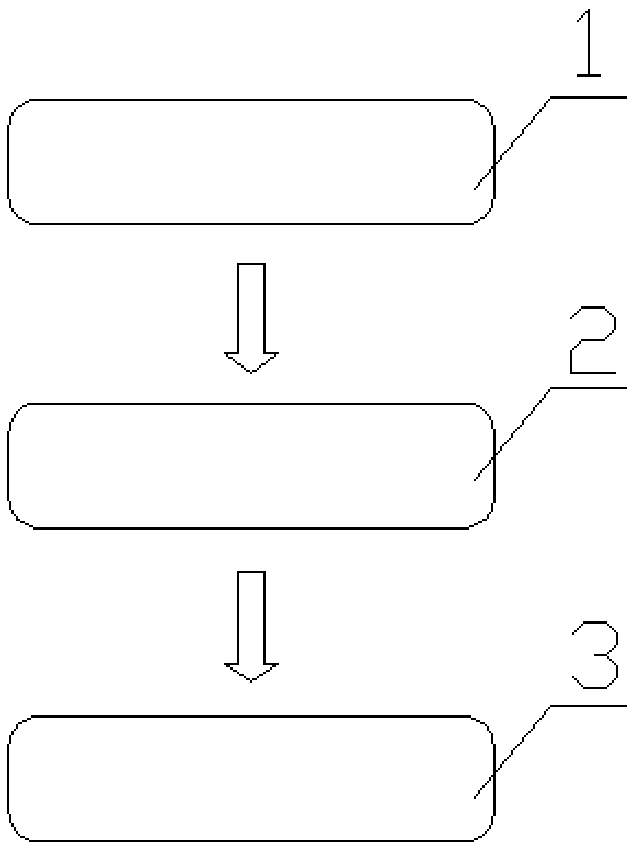
[0027] see figure 1 Shown, a system for analyzing patterns of association change from multi-omics data comprising:
[0028] Omics dataset 1 containing various omics data;
[0029] Binarization algorithm unit 2 for preprocessing omics datasets;
[0030] Association rule Apriori algorithm unit 3 for mining out the interrelated patterns of molecular changes.
[0031] In this embodiment, the omics data set includes genome exon sequencing data, genome copy number change data, genome methylation data, gene chip expression profile data, gene sequencing data, non-coding RNA expression data, and protein profile data . It should be noted that the omics data set of the present invention is not limited to include the above-mentioned types of data, and can be increased according to actual needs.
[0032] A method for analyzing patterns of association change from multi-omics data, comprising the steps of:
[0033] Step 1. Collecting and arranging various omics data to form an omics dat...
Embodiment 2
[0043] Utilizing the RNA-seq omics data of patients with esophageal cancer, a total of 14,179 interrelated rules between genes are excavated by the present invention. For example, one of the rules is {CDK1}==>{CCNB2}, the support is 0.435, the confidence is 0.808, and the lift is 1.416. It shows that CDK1 (cyclin dependent kinase 1, cyclin dependent kinase 1) and CCNB2 (cyclin B2, cyclin B2) have a high correlation. In the RNA-seq gene detection data of 43.5% of esophageal cancer patients, it was found that the two were differentially expressed at the same time; when CDK1 was differentially expressed, the probability of CCNB2 was also found to be differentially expressed as high as 80.8%; the probability of both differentially expressed at the same time was CDK1 was 1.416 times more likely to be differentially expressed independently of CCNB2.
Embodiment 3
[0045] In order to study the similarity of DNA methylation mechanism between esophageal cancer and breast cancer, we used the present invention to analyze the DNA methylomics data of esophageal cancer patients and breast cancer patients respectively, and then compared the results of the two. When the support degree is 0.4 and the confidence degree is 0.8, a total of 239 common association rules are found. We found that many of these rule-forming genes are cadherins, which play a role in cell adhesion. This result shows that the methylation of cell adhesion-related genes is a common carcinogenic mechanism of esophageal cancer and breast cancer.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com