Deep learning method for predicting prognosis risk of cancer patient based on multi-omics data
A technology of omics data and deep learning, applied in machine learning, medical informatics, informatics, etc., can solve problems such as inability to solve target data sets and low accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
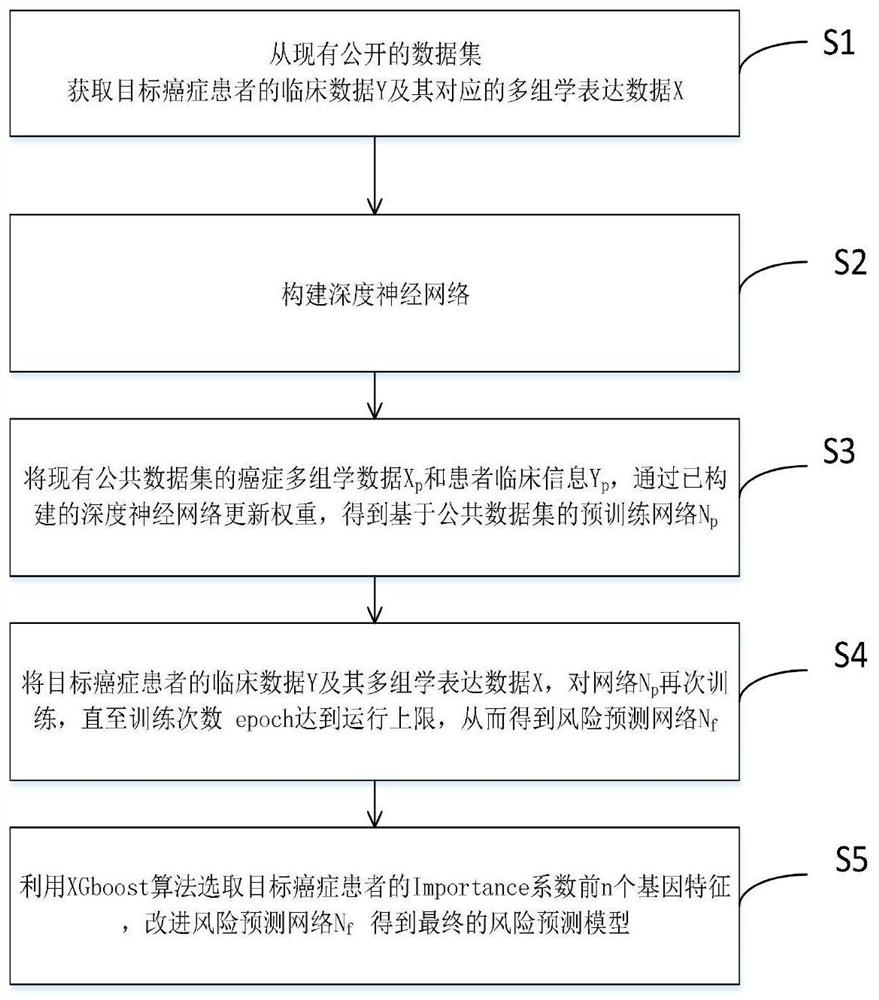
[0044] Such as figure 1 As shown, a deep learning method for predicting the prognosis risk of cancer patients based on multi-omics data is used to predict the prognosis risk of cancer patients, including the following steps:
[0045] S1: Obtain clinical data Y and corresponding multi-omics expression data X of target cancer patients from existing public data sets (such as TCGA, GEO);
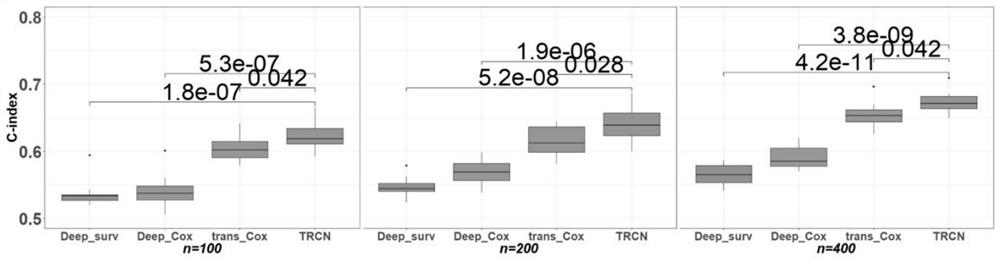
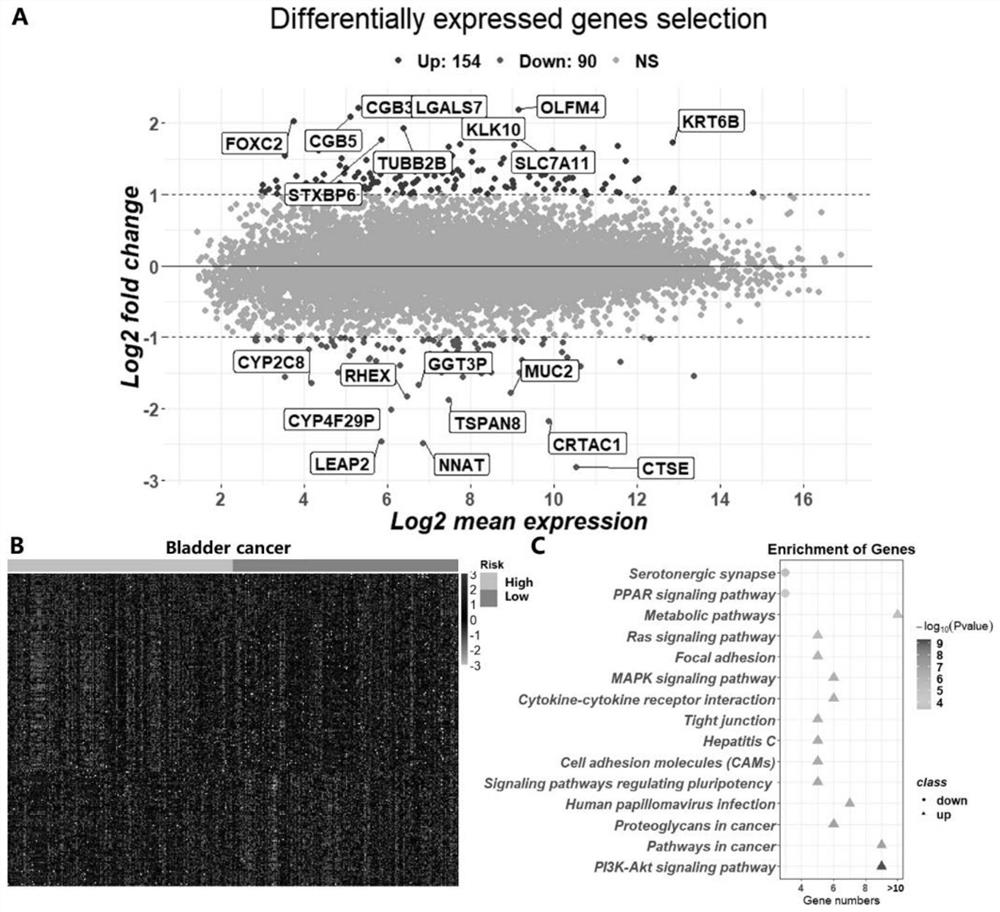
[0046] In a specific embodiment, 14 TCGA data sets (BRCA, CESC, COAD, ESCA, HNSC, KIRC, LGG, LIHC, LUAD, LUSC, MESO, PAAD, SRAC and SKCM) were used for pre-training, and bladder cancer ( BLCA) data as the target cancer.
[0047] Among them, multi-omics data includes mRNA expression, miRNA expression, DNA methylation information and copy number variation information of bladder cancer patients. mRNA data is RNA sequencing data generated by UNC Illumina HiSeq_RNASeq V2. miRNA is the miRNA sequencing data obtained by BCGSC Illumina HiSeq miRNASeq. DNA methylation data was generated by USCHumanMe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com