Patents
Literature
3131results about "Biological models" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
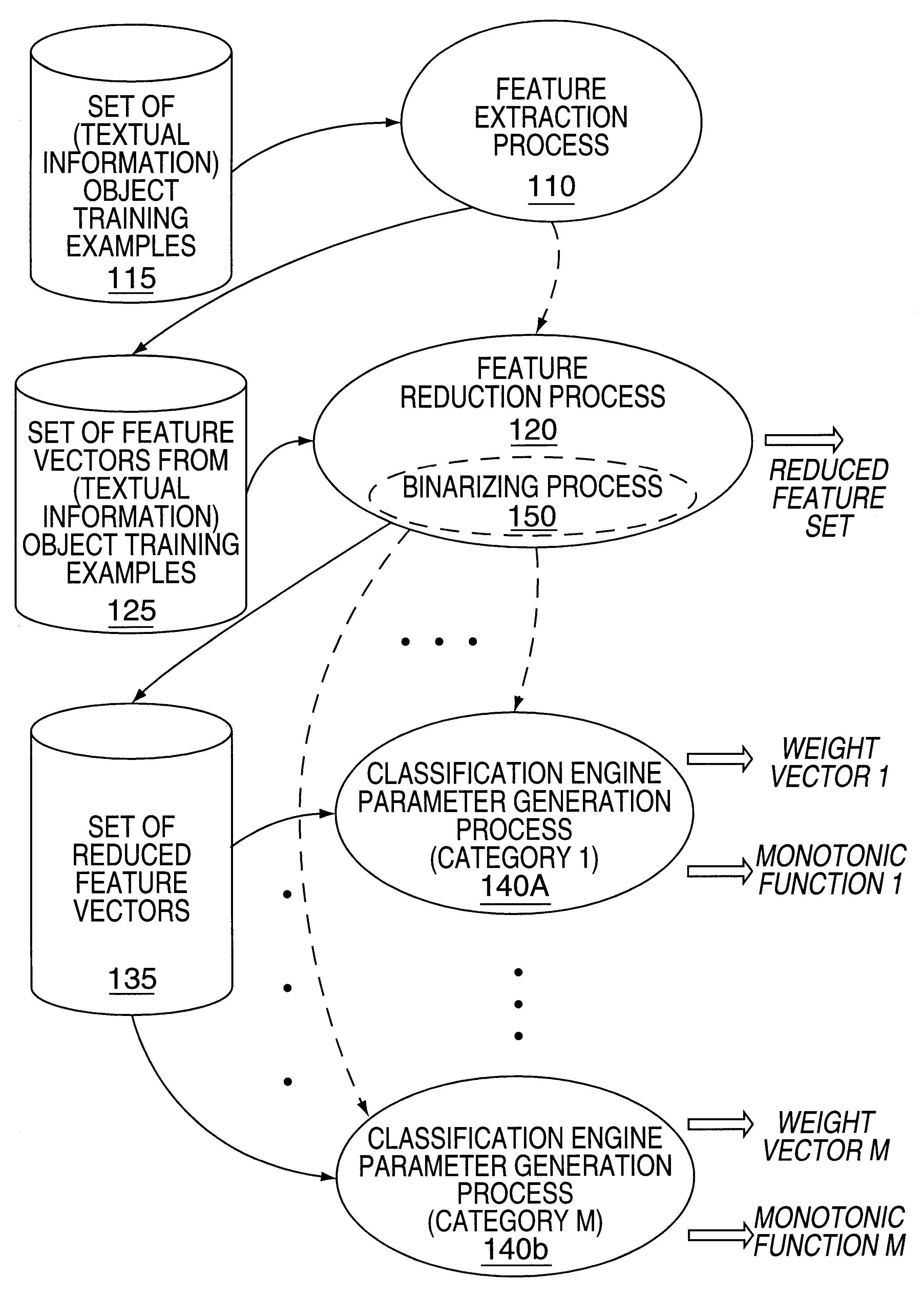
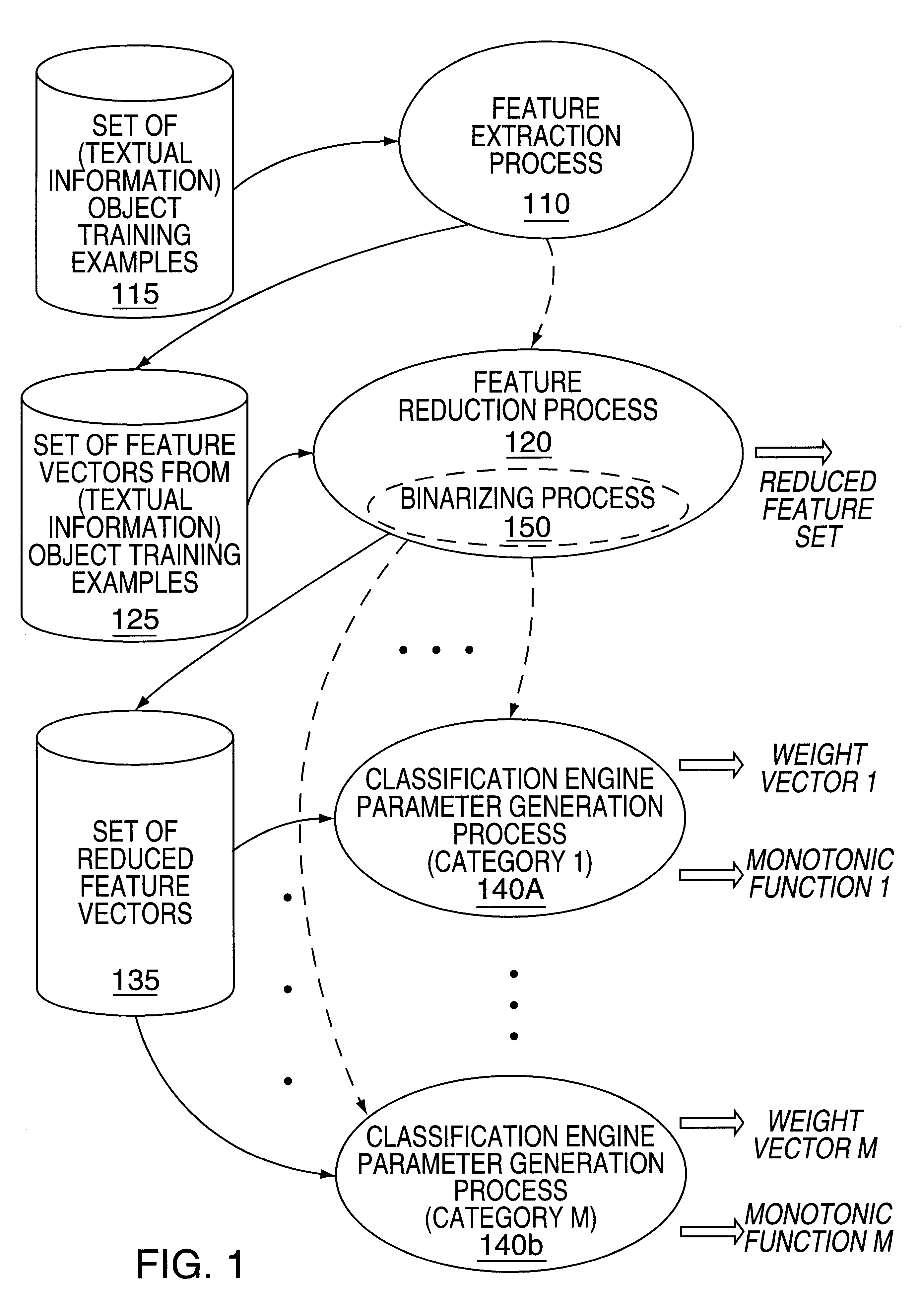
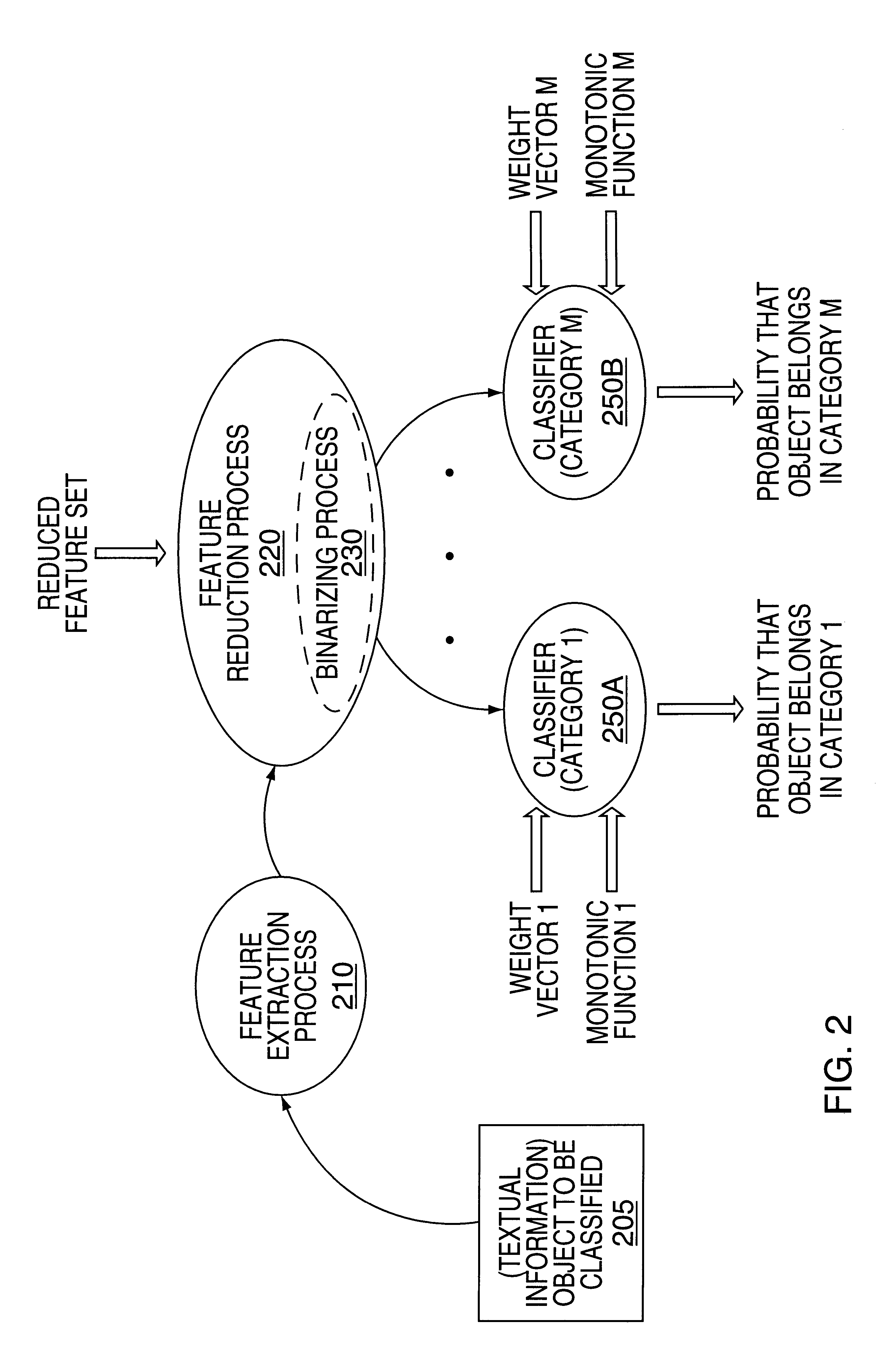
Methods and apparatus for classifying text and for building a text classifier
InactiveUS6192360B1Quick calculationData processing applicationsDigital data information retrievalPattern recognitionText categorization
Owner:MICROSOFT TECH LICENSING LLC
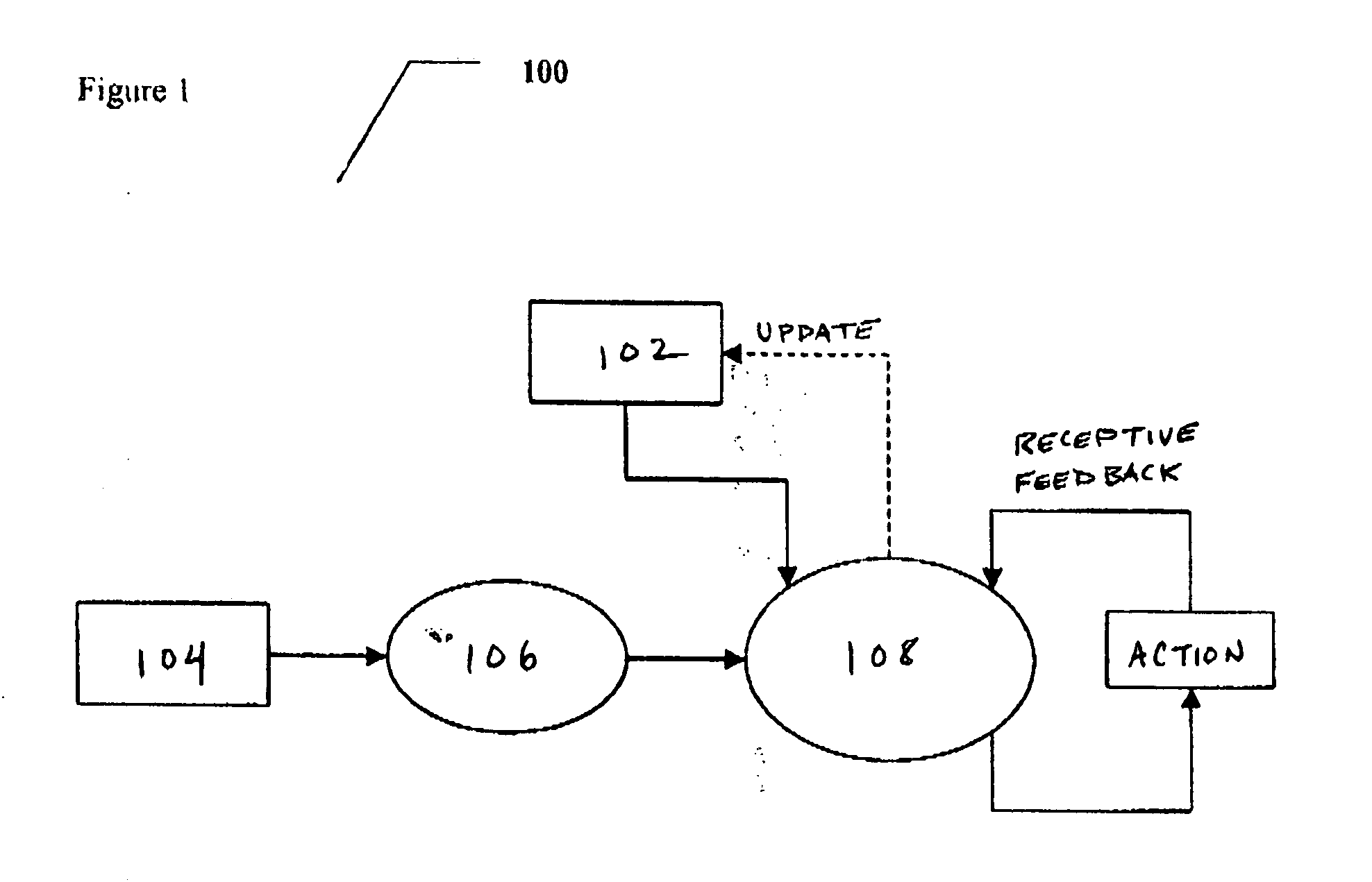
Proactive user interface
InactiveUS20050054381A1Increase sense of intelligenceImprove experienceInput/output for user-computer interactionDevices with sensorDisplay deviceTouchscreen
A proactive user interface, which could optionally be installed in (or otherwise control and / or be associated with) any type of computational device. The proactive user interface actively makes suggestions to the user, based upon prior experience with a particular user and / or various preprogrammed patterns from which the computational device could select, depending upon user behavior. These suggestions could optionally be made by altering the appearance of at least a portion of the display, for example by changing a menu or a portion thereof; providing different menus for display; and / or altering touch screen functionality. The suggestions could also optionally be made audibly.
Owner:SAMSUNG ELECTRONICS CO LTD
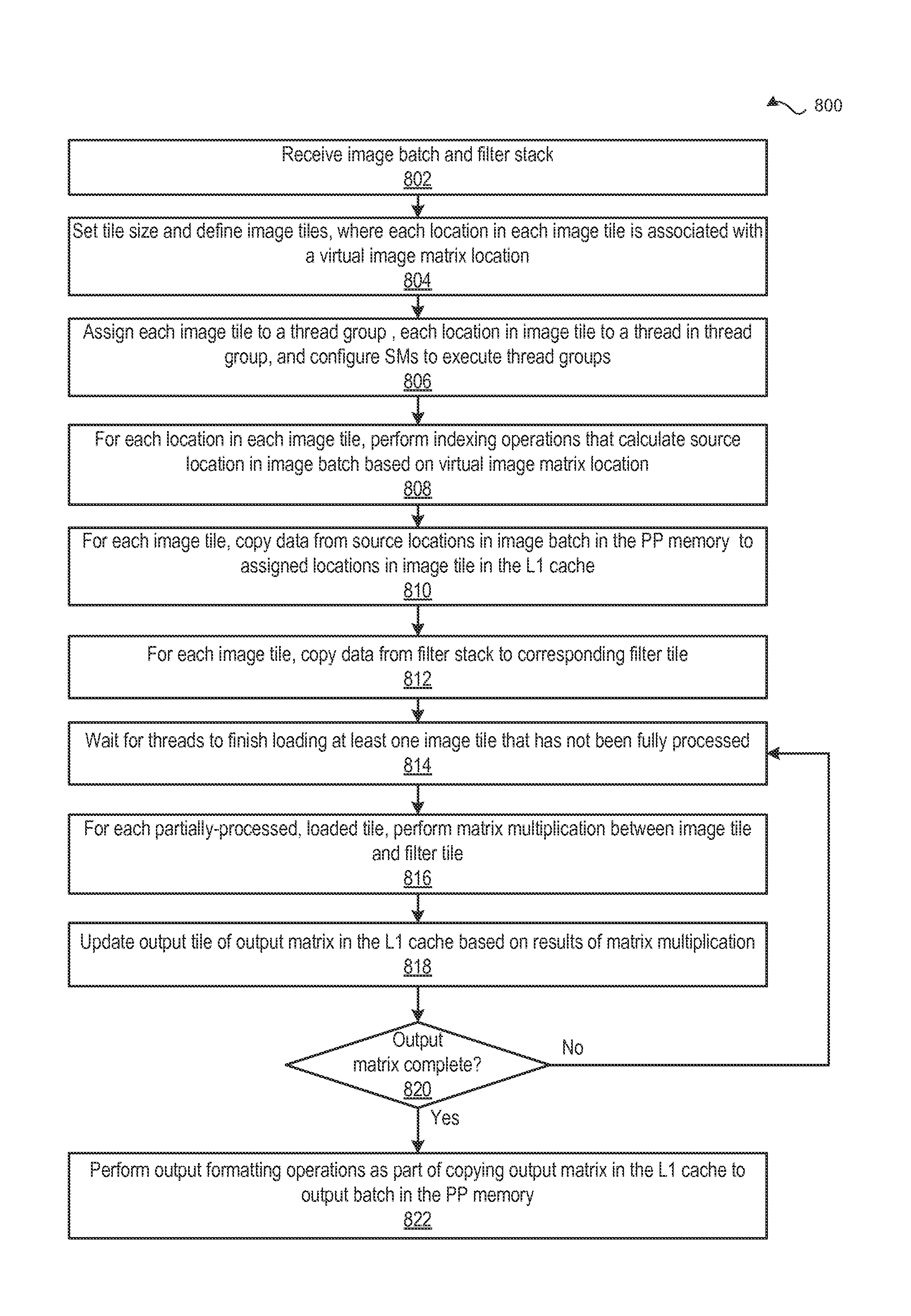
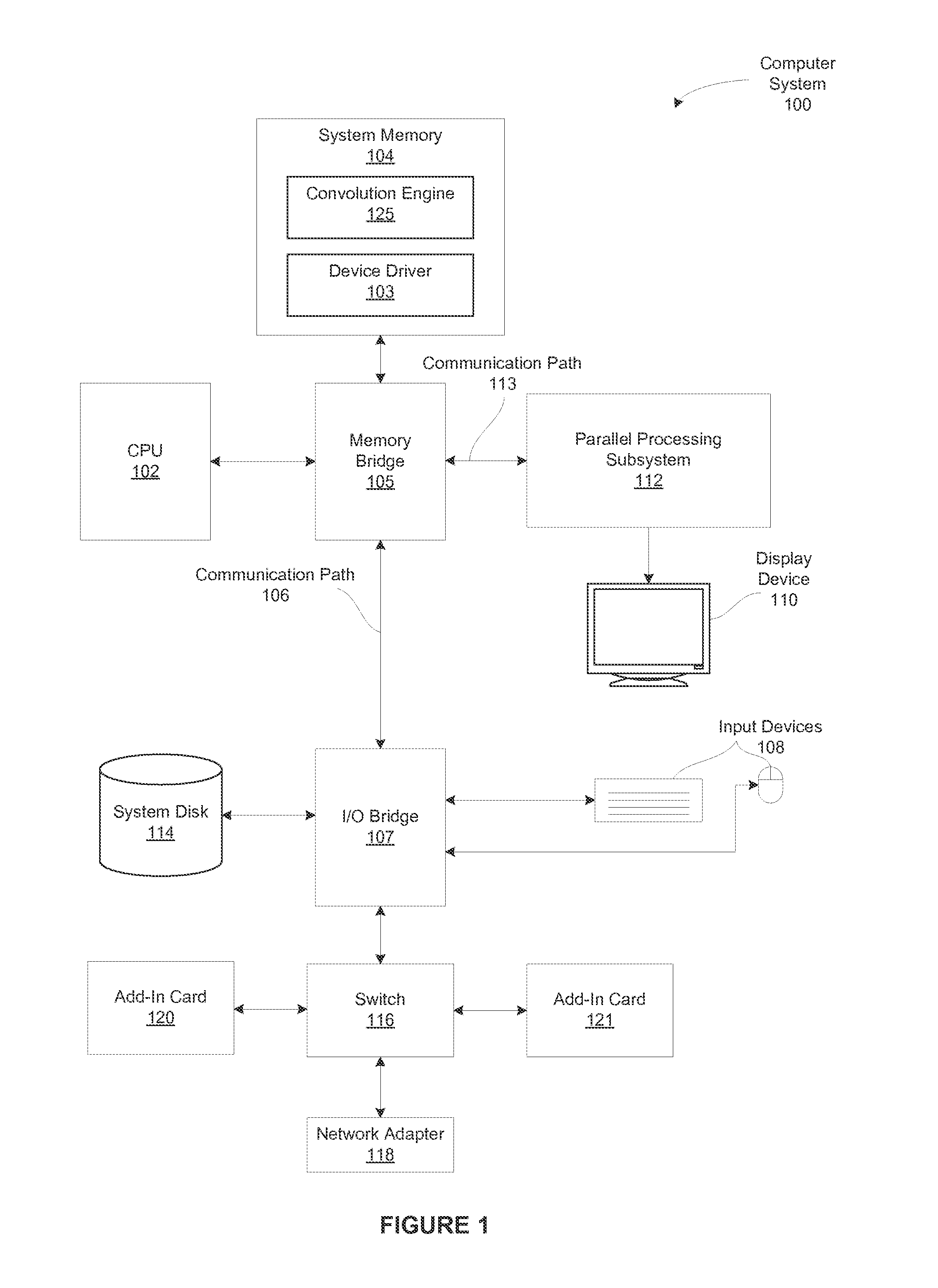
Performing multi-convolution operations in a parallel processing system
ActiveUS20160062947A1Easy to operateOptimizing on-chip memory usageBiological modelsComplex mathematical operationsLine tubingParallel processing
In one embodiment of the present invention a convolution engine configures a parallel processing pipeline to perform multi-convolution operations. More specifically, the convolution engine configures the parallel processing pipeline to independently generate and process individual image tiles. In operation, for each image tile, the pipeline calculates source locations included in an input image batch. Notably, the source locations reflect the contribution of the image tile to an output tile of an output matrix—the result of the multi-convolution operation. Subsequently, the pipeline copies data from the source locations to the image tile. Similarly, the pipeline copies data from a filter stack to a filter tile. The pipeline then performs matrix multiplication operations between the image tile and the filter tile to generate data included in the corresponding output tile. To optimize both on-chip memory usage and execution time, the pipeline creates each image tile in on-chip memory as-needed.
Owner:NVIDIA CORP
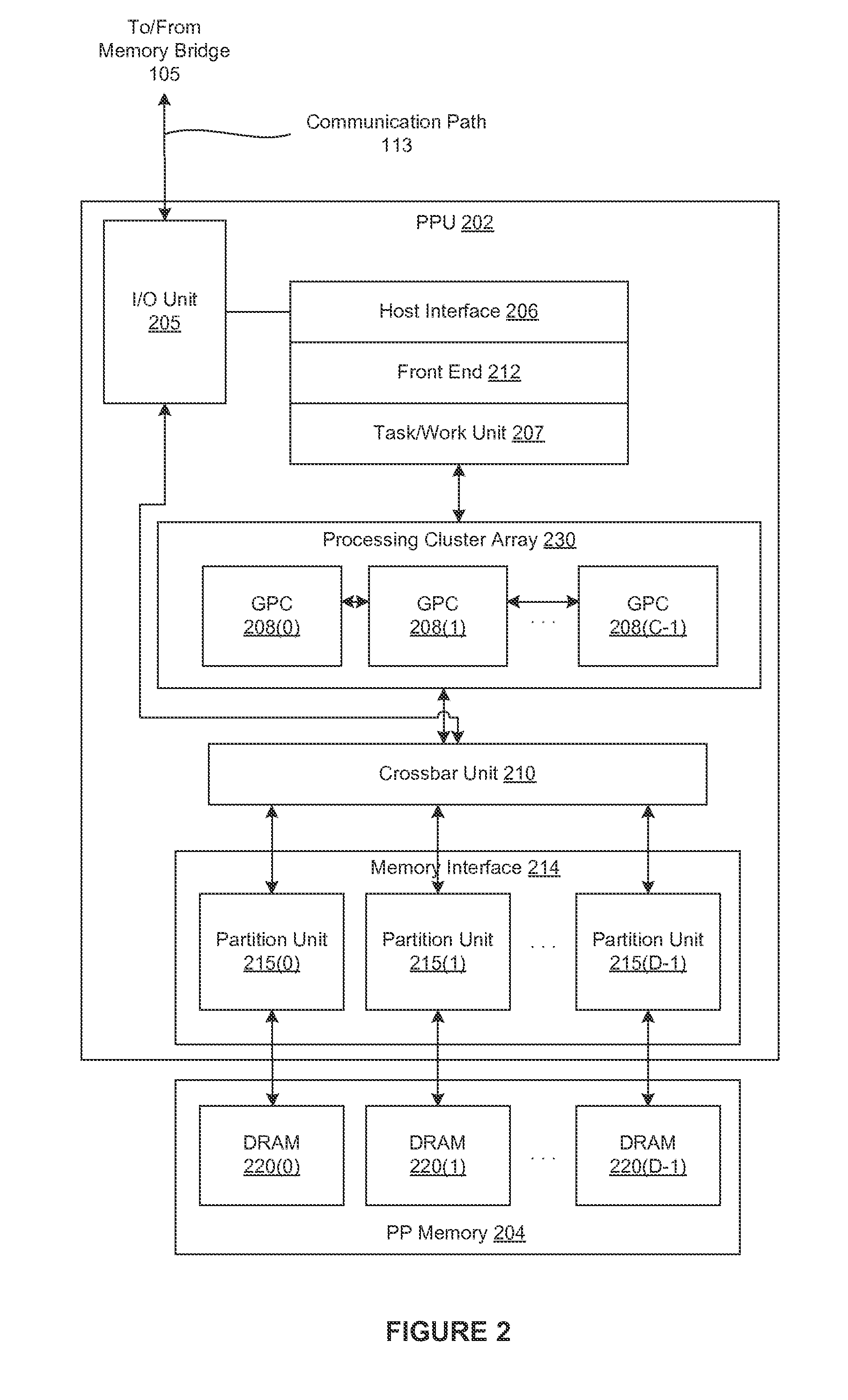
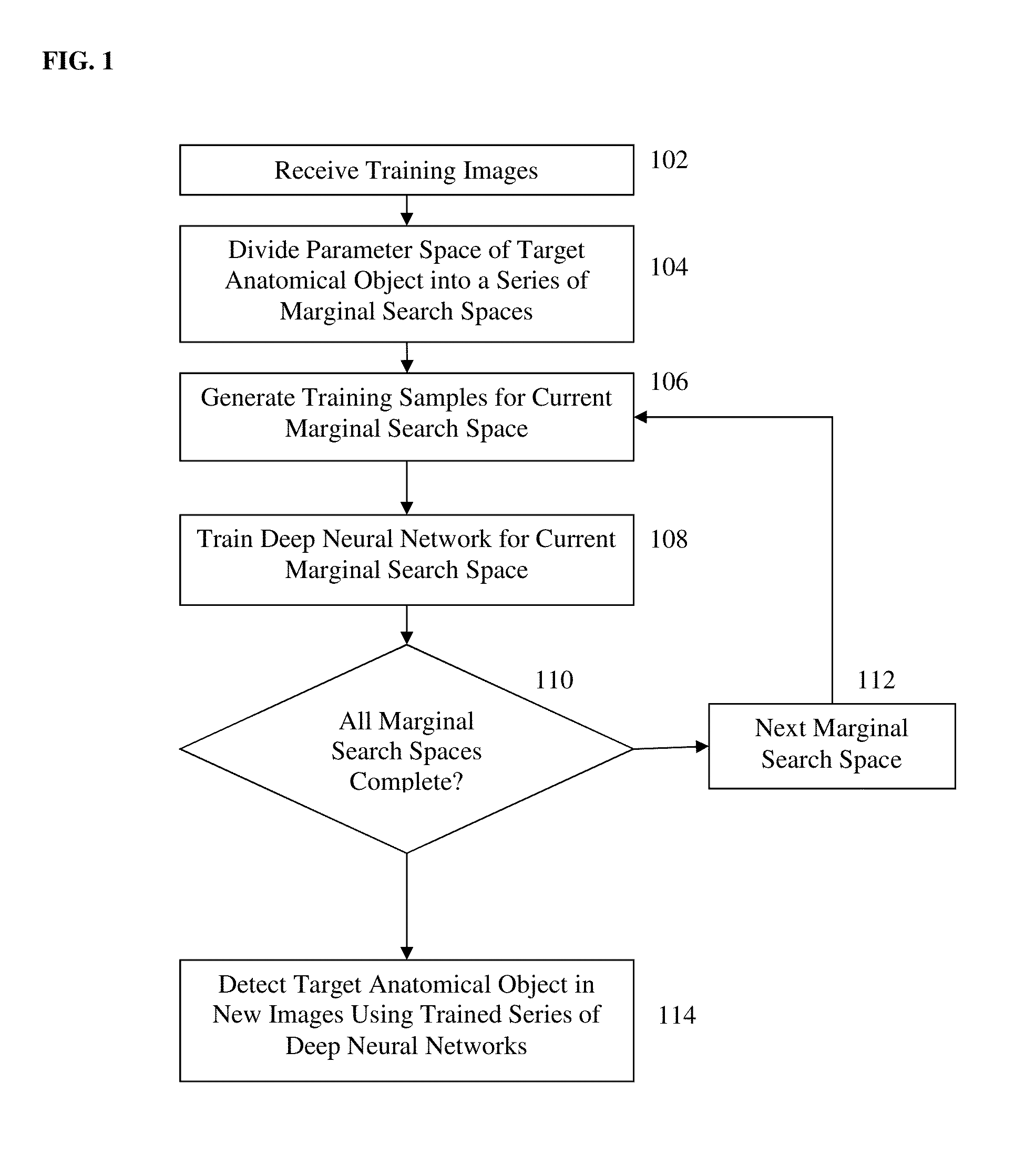
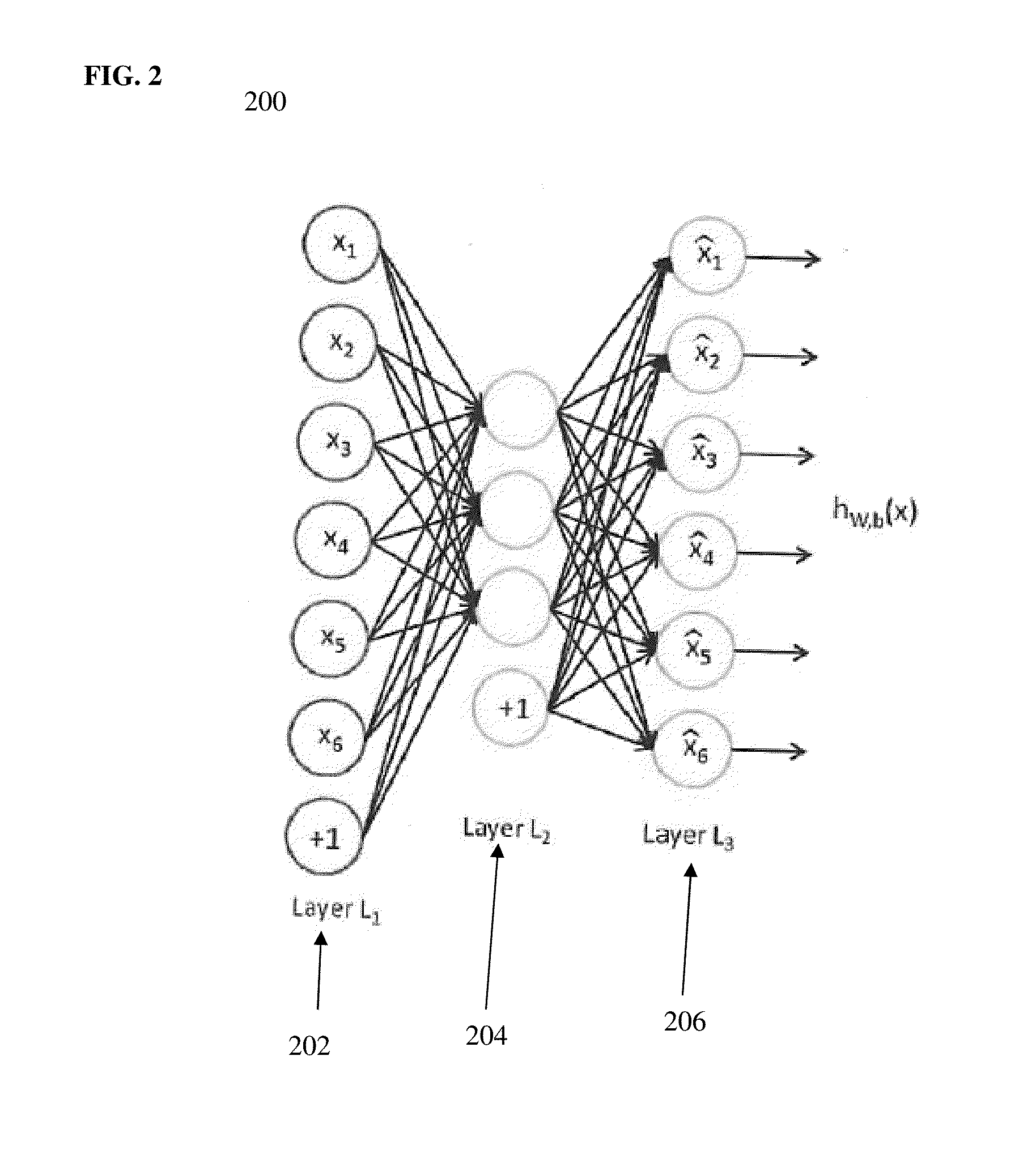
Method and System for Anatomical Object Detection Using Marginal Space Deep Neural Networks
ActiveUS20160174902A1Add dimensionUltrasonic/sonic/infrasonic diagnosticsImage enhancementMachine learningObject detection
A method and system for anatomical object detection using marginal space deep neural networks is disclosed. The pose parameter space for an anatomical object is divided into a series of marginal search spaces with increasing dimensionality. A respective sparse deep neural network is trained for each of the marginal search spaces, resulting in a series of trained sparse deep neural networks. Each of the trained sparse deep neural networks is trained by injecting sparsity into a deep neural network by removing filter weights of the deep neural network.
Owner:SIEMENS HEALTHCARE GMBH
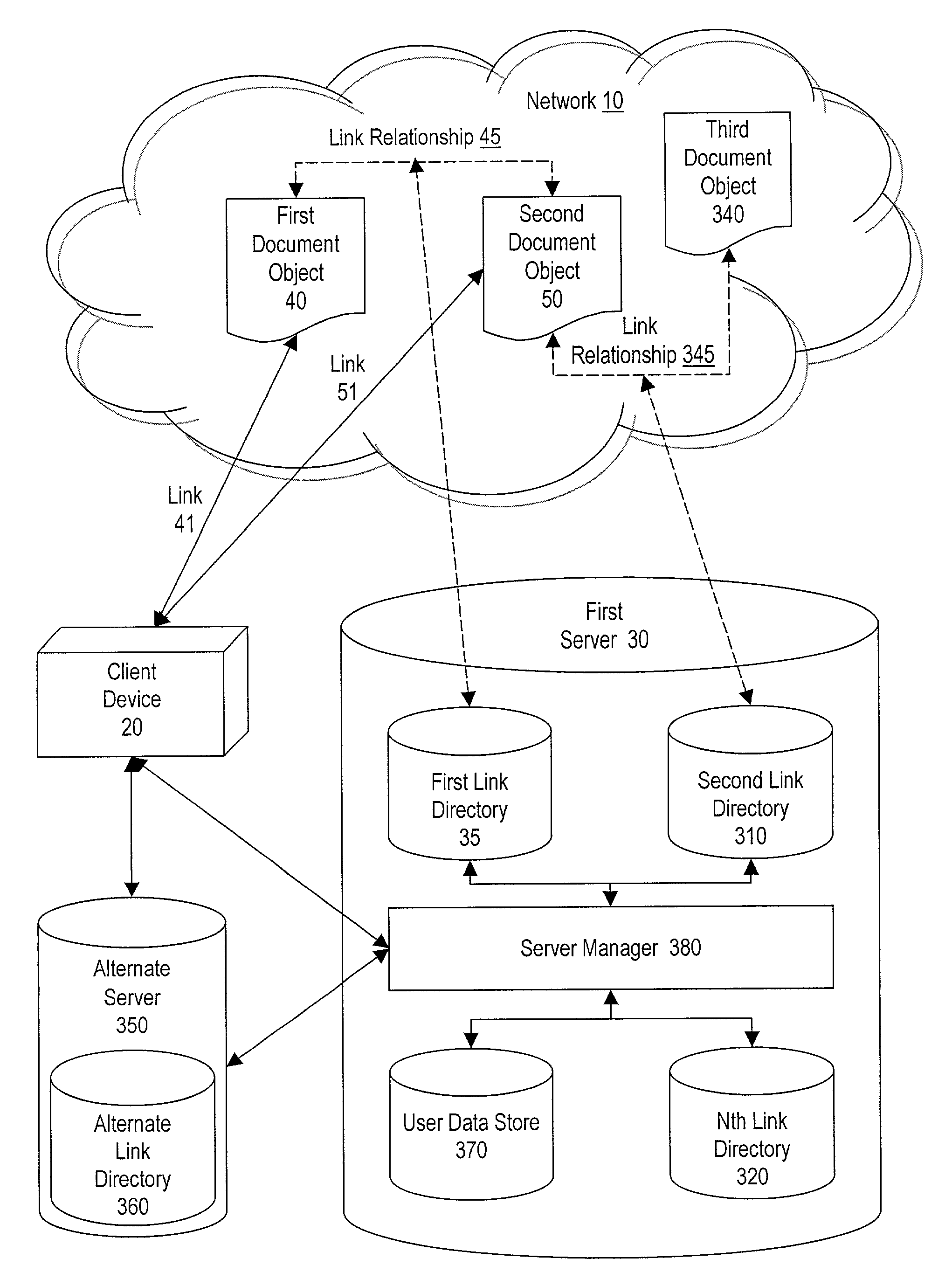
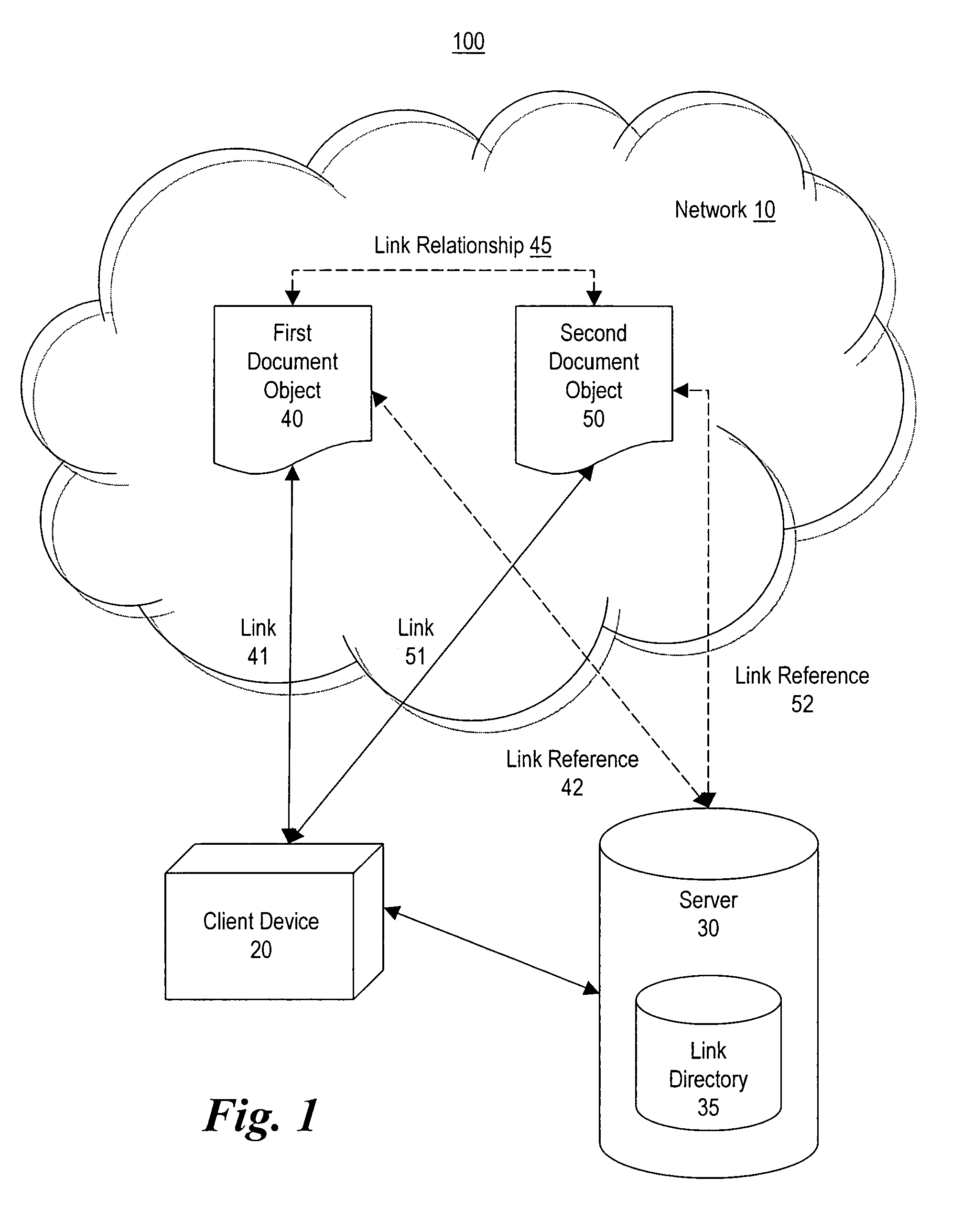
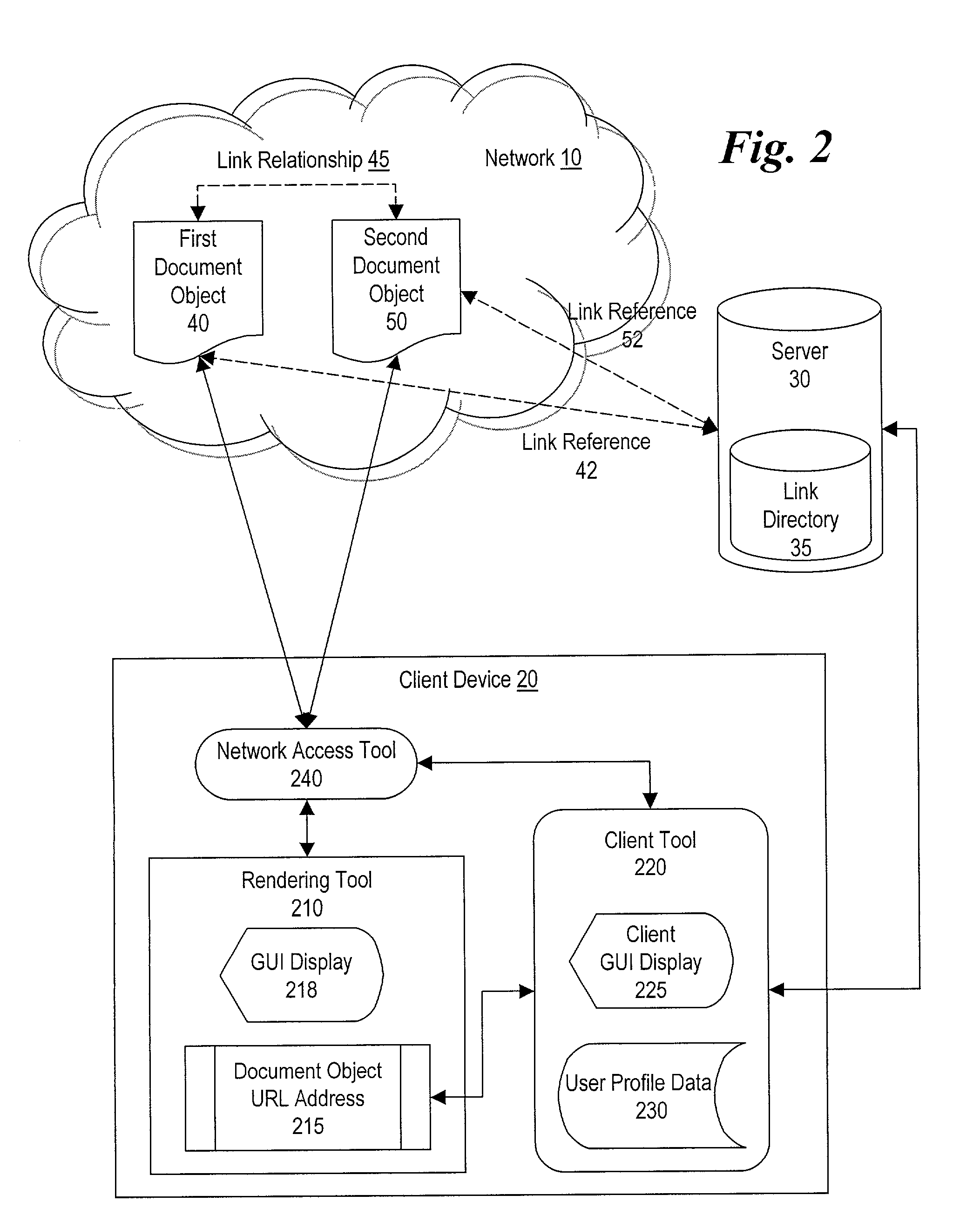
Method and system for making document objects available to users of a network
InactiveUS7111232B1Improve browsing experienceBiological modelsWebsite content managementDatabaseLink relation
Owner:DSS TECH MANAGEMENT
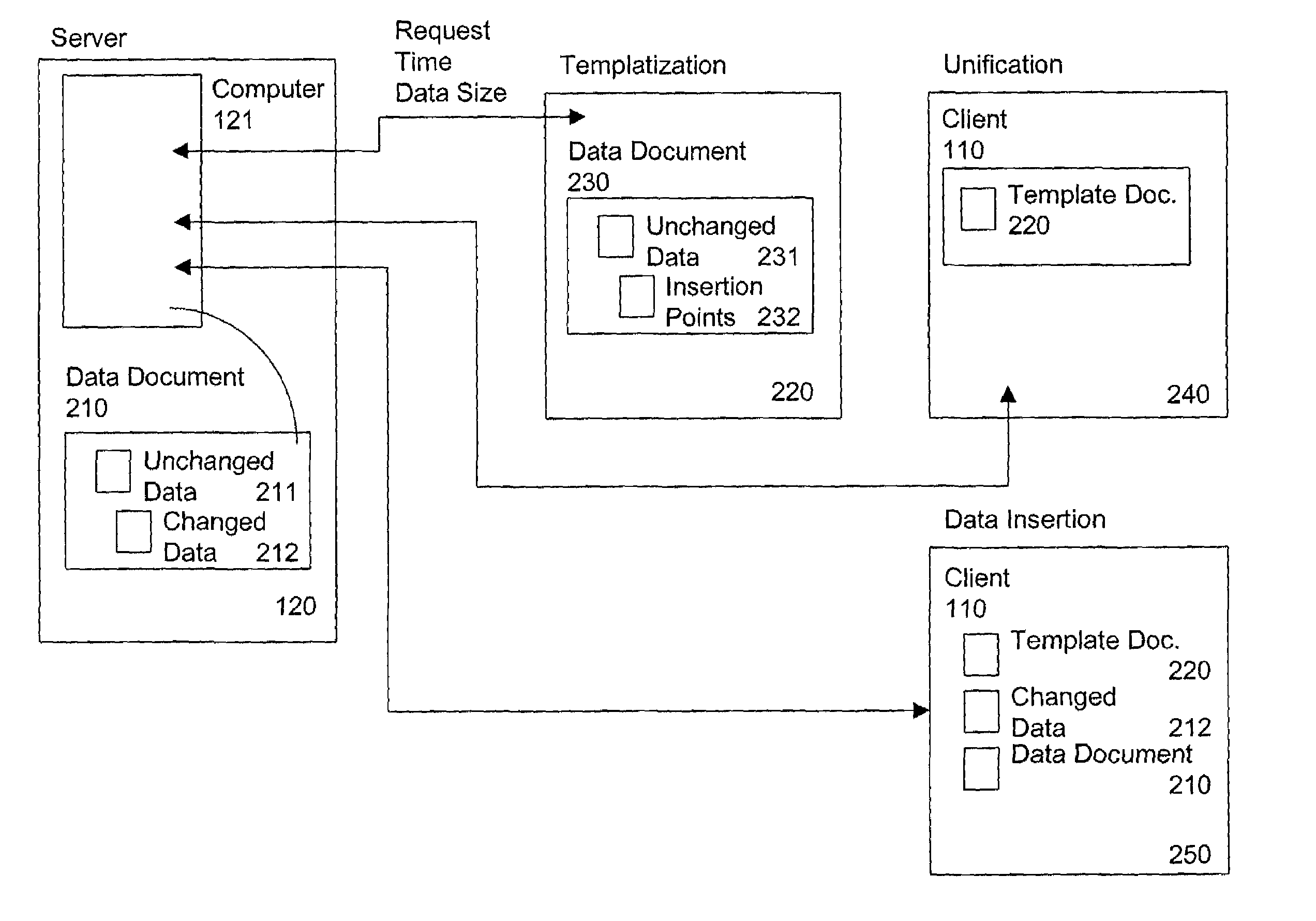
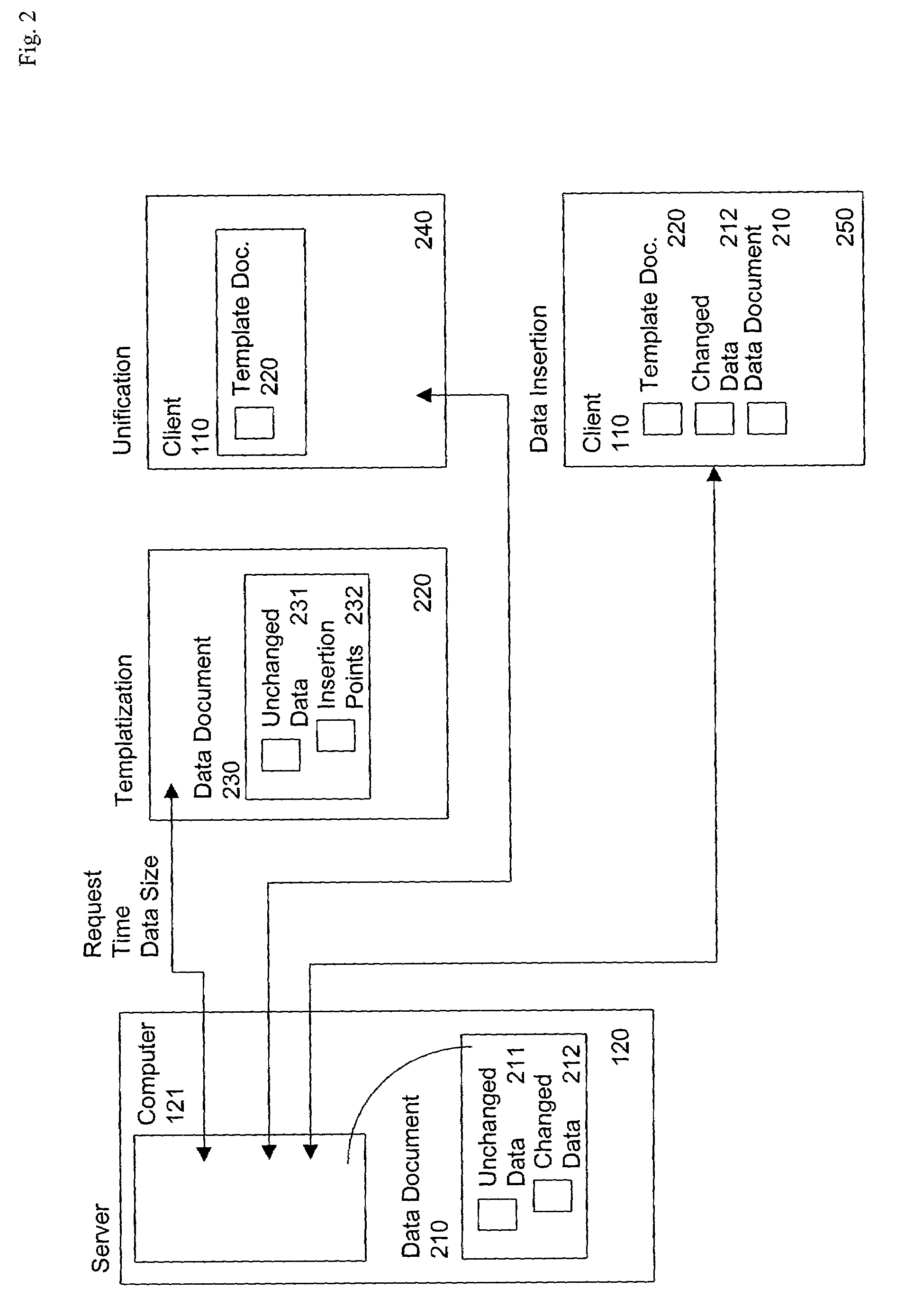
Server-originated differential caching
InactiveUS7269784B1Minimize timeReduce the amount requiredDigital data information retrievalBiological modelsUnique identifierClient-side
The invention provides a method and system for sending relatively identical web pages, when requested by subsequent users, with substantial reduction of bandwidth. The server determines a “template web page” corresponding to the actual information on the web page, and having a set of insertion points, at which changed data can be inserted by the client. The server sends a JavaScript program corresponding to the template web page, which makes reference to the template web page and the changed data, along with sending the actual changed data itself. A first user requesting the web page receives the entire web page, while a second user requesting the web page (or the first user re-requesting the web page at a later time) receives the template information plus only the changed data. The server re-determines the template web page from time to time, such as when a ratio of changed data to template web page data exceeds a selected threshold. The server identifies the particular template web page to the client using a unique identifier (an “E-tag”) for the particular data sent in response to the request. Since the E-tag refers to the template, not the underlying web page, when the standard client makes its conditional request for the web page “if not changed”, the server responds that the web page is “not changed” even if it really is, but embeds the changed data in a cookie it sends to the client with the server response to the client request.
Owner:DIGITAL RIVER INC
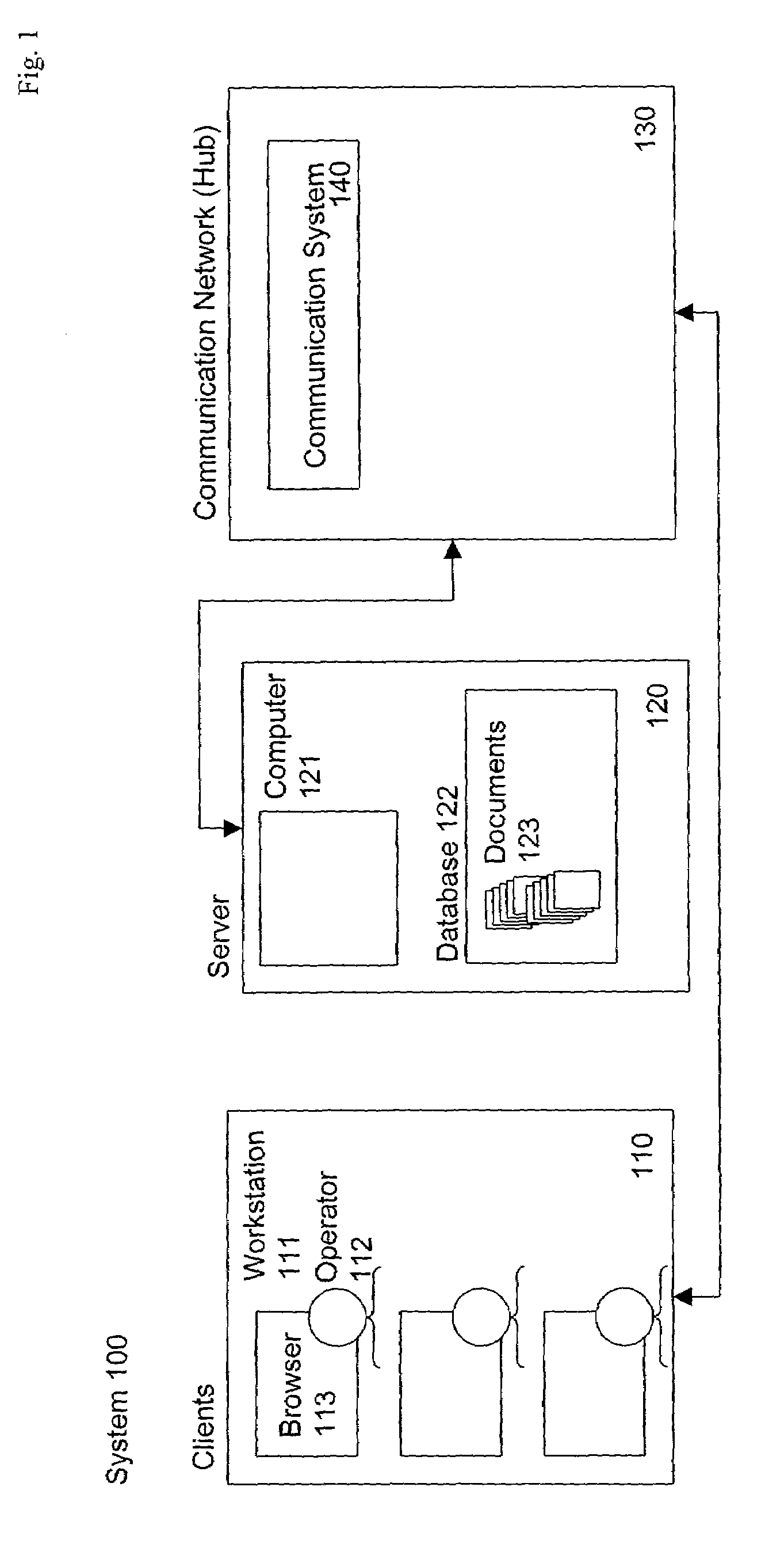
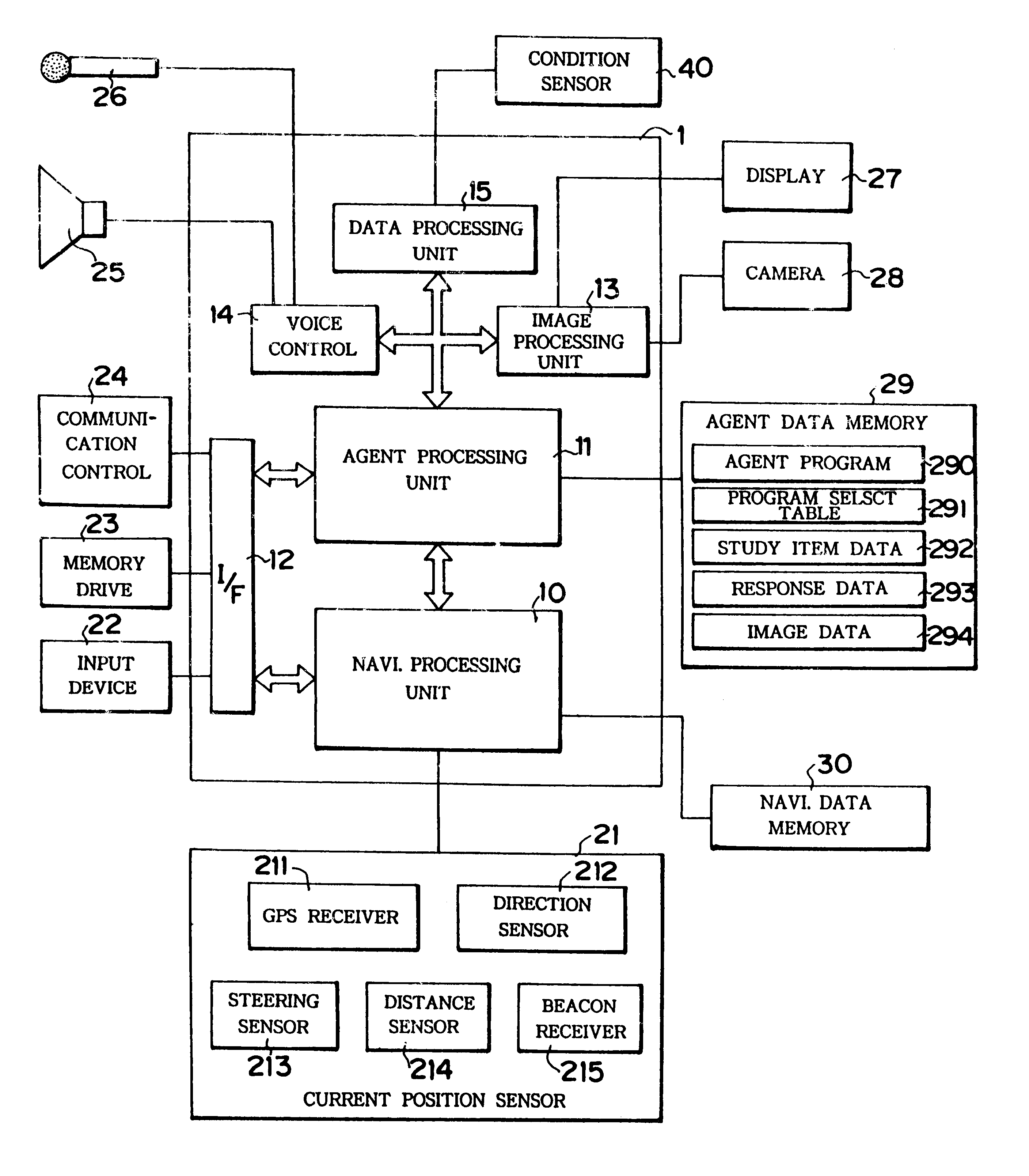
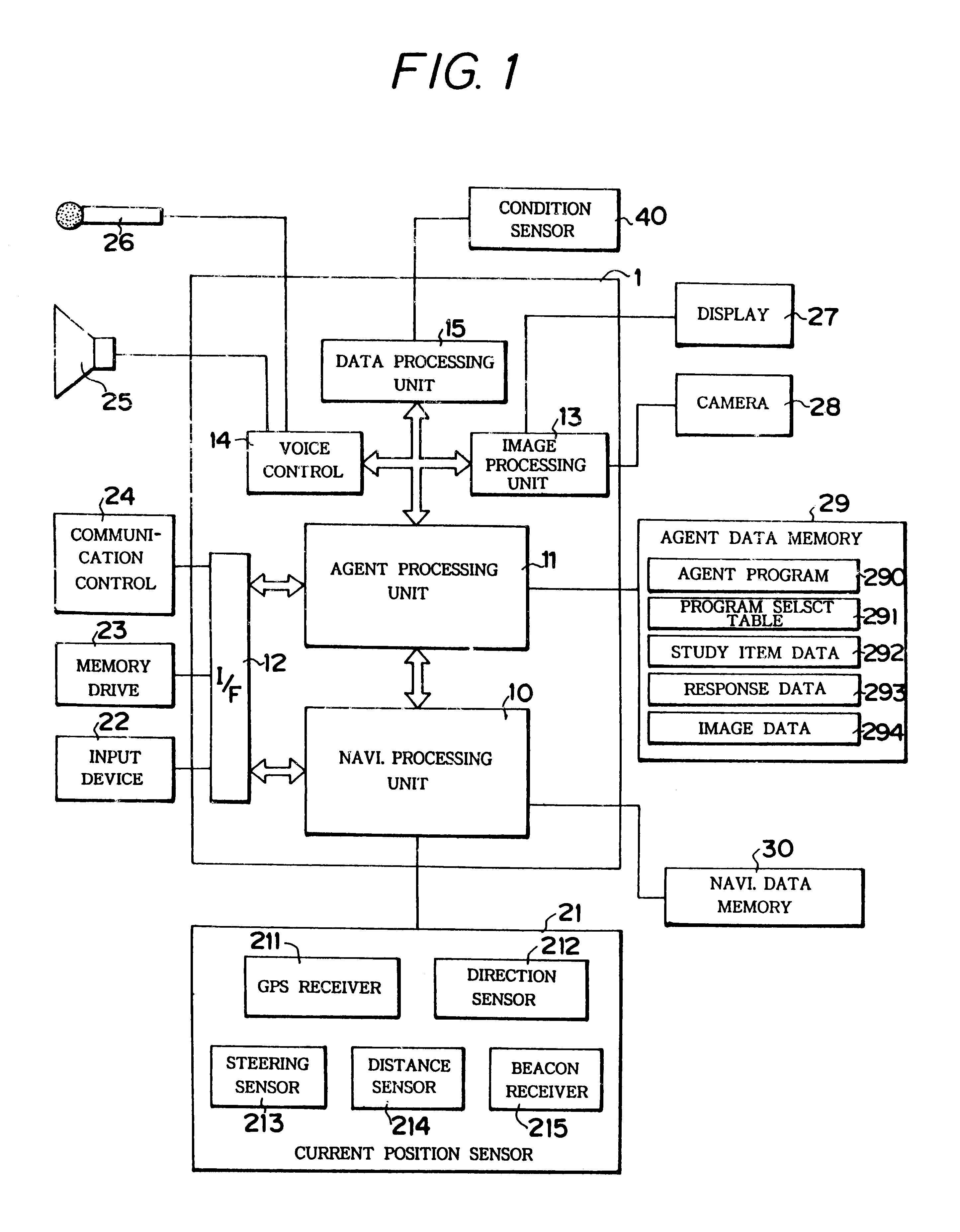
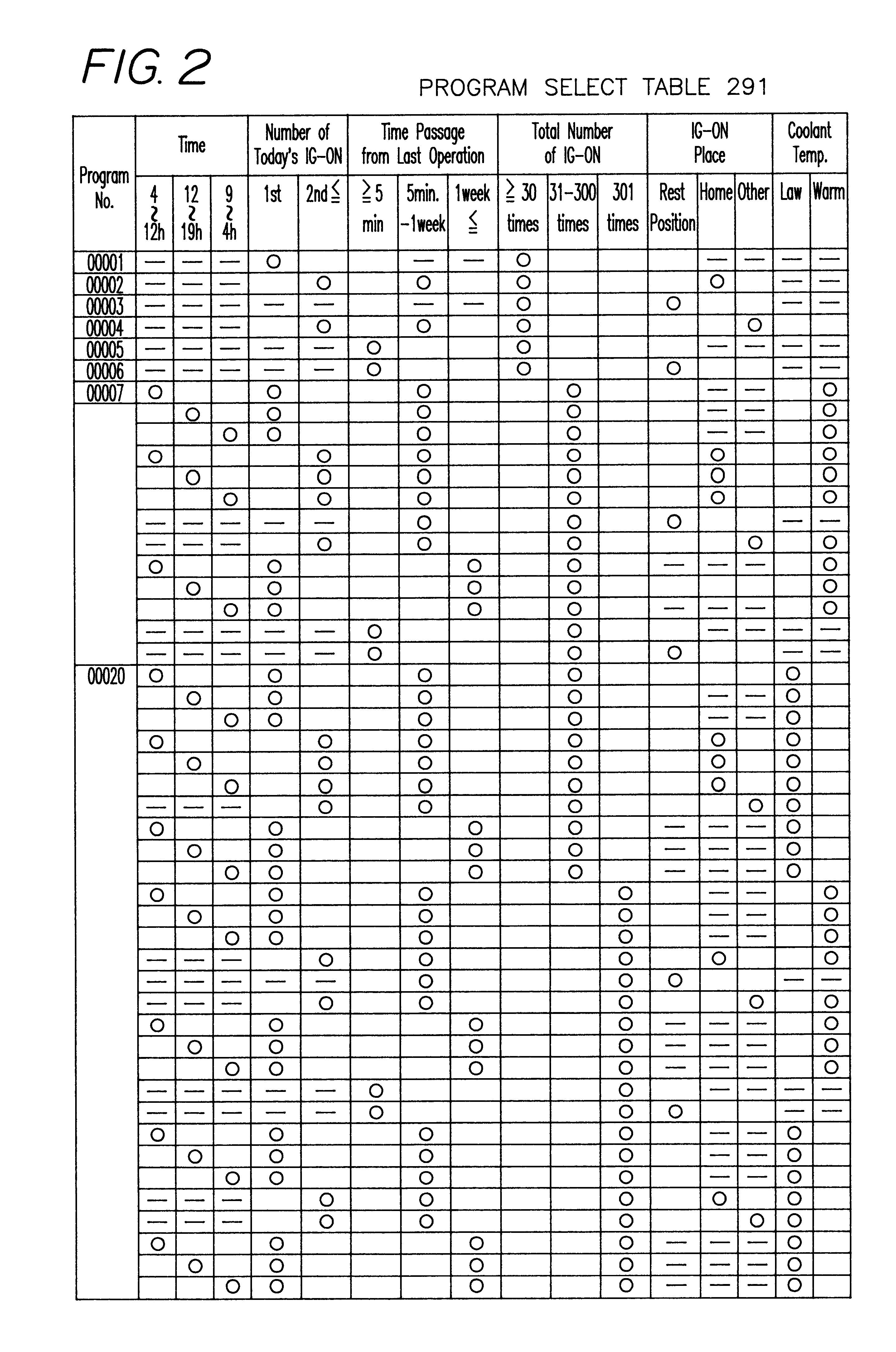
Device mounted in vehicle
InactiveUS6249720B1Instruments for road network navigationLighting and heating apparatusDriver/operatorIn vehicle
A device is mounted in a vehicle which allows one or more of personified agent or imaginary living body to appear before a driver or passengers in the vehicle for communication therewith. A current status sensor (40) detects predetermined vehicle current status data, and a memory (292, 293) stores study data in response to entry of the latest current status data. One of a plurality of communication programs stored in a program table (291) may be selected in accordance with the current status data and / or the study data. An agent control unit (11) controls activity of the agent in accordance with the selected communication program. The determined agent activity is outputted through a display (27) and a speaker (25) for communication with the driver.
Owner:EQUOS RES
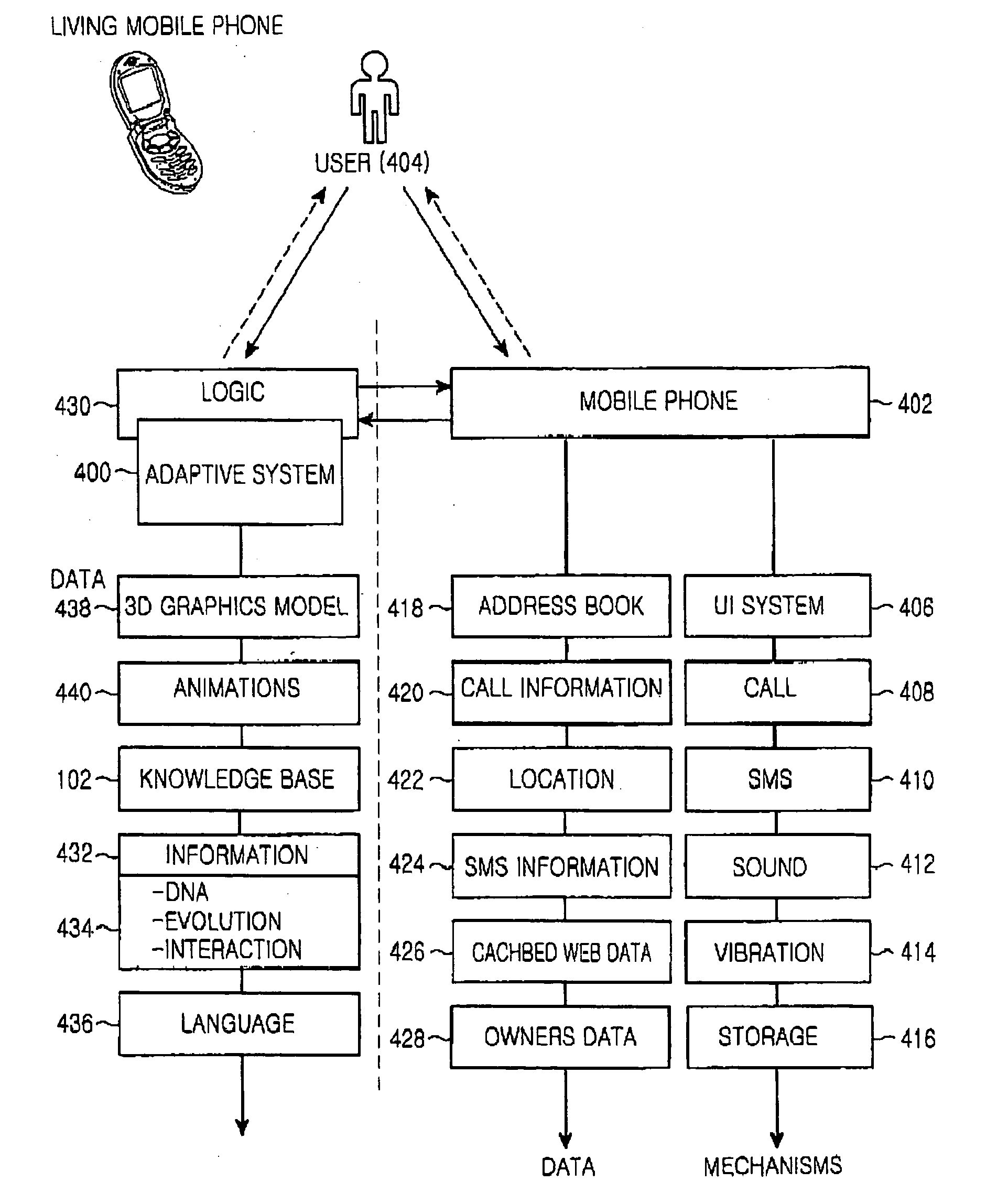
Proactive user interface including evolving agent
ActiveUS20050118996A1Enhanced interactionImprove user experienceDevices with sensorDigital computer detailsDisplay deviceTouchscreen
A proactive user interface, installed in (or otherwise control and / or be associated with) any type of computational device. The proactive user interface actively makes suggestions to the user, based upon prior experience with a particular user and / or various preprogrammed patterns from which the computational device could select, depending upon user behavior. These suggestions can be made by altering the appearance of at least a portion of the display, for example by changing a menu or a portion thereof; providing different menus for display; and / or altering touch screen functionality. The suggestions can also be made audibly.
Owner:SAMSUNG ELECTRONICS CO LTD
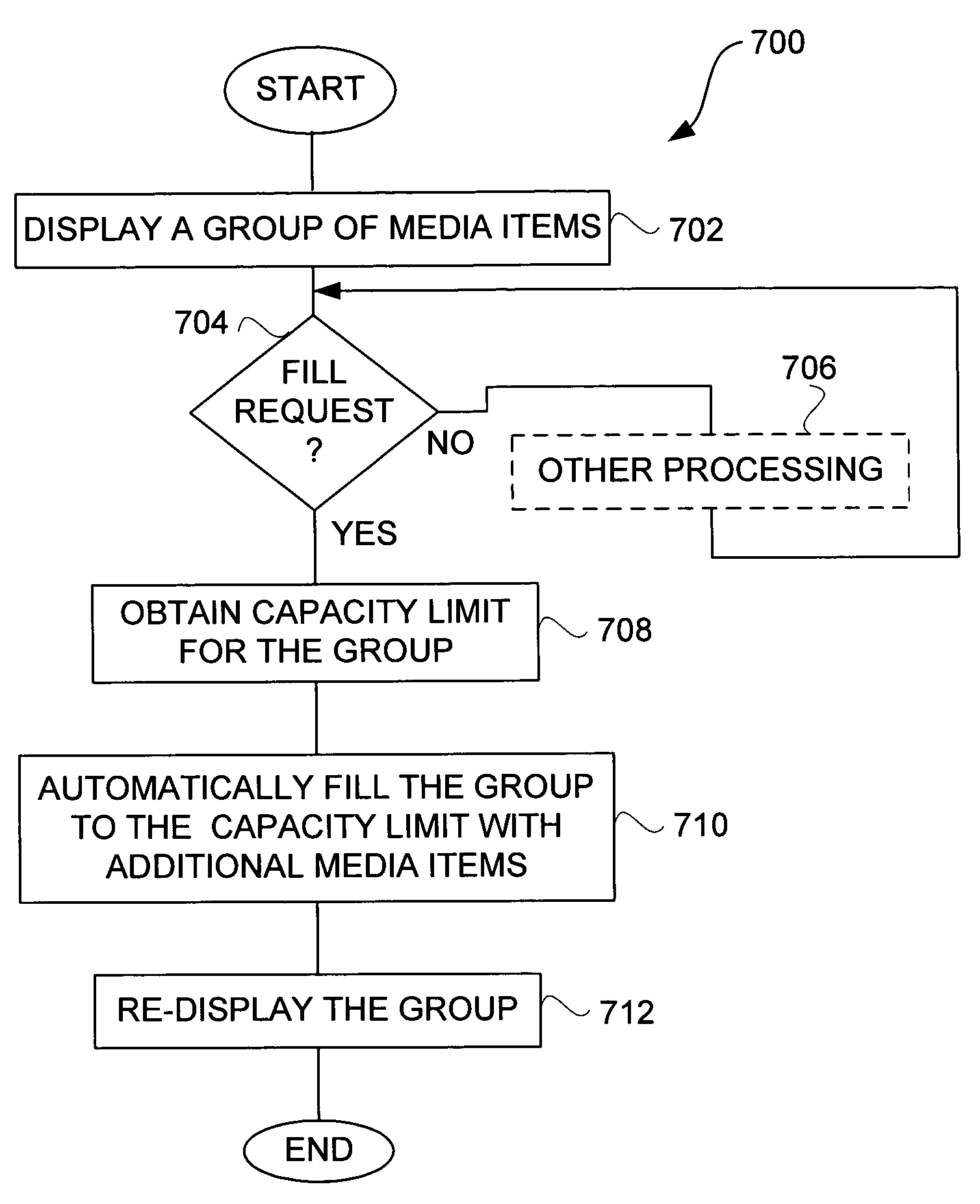
Media management for groups of media items
Improved techniques to utilize and manage a group of media items (or media assets) on a computing device are disclosed. The group of media items can be utilized and managed at a host computer for the host computer as well as a media device (e.g., media player) that can couple to the host computer. One popular example of a group of media items is know as a playlist, which can pertain to a group of audio tracks. One aspect pertains to a graphical user interface that enables a user to trade-off storage capacity of a media device between media asset storage and data storage. Another aspect pertains to a graphical user interface that assists a user with selecting media items to fill a group of media items. Still another aspect pertains to providing a persistent media device playlist at a host computer. Yet still another aspect pertains to imposing capacity limits to a playlist, such as a media device playlist.
Owner:APPLE INC
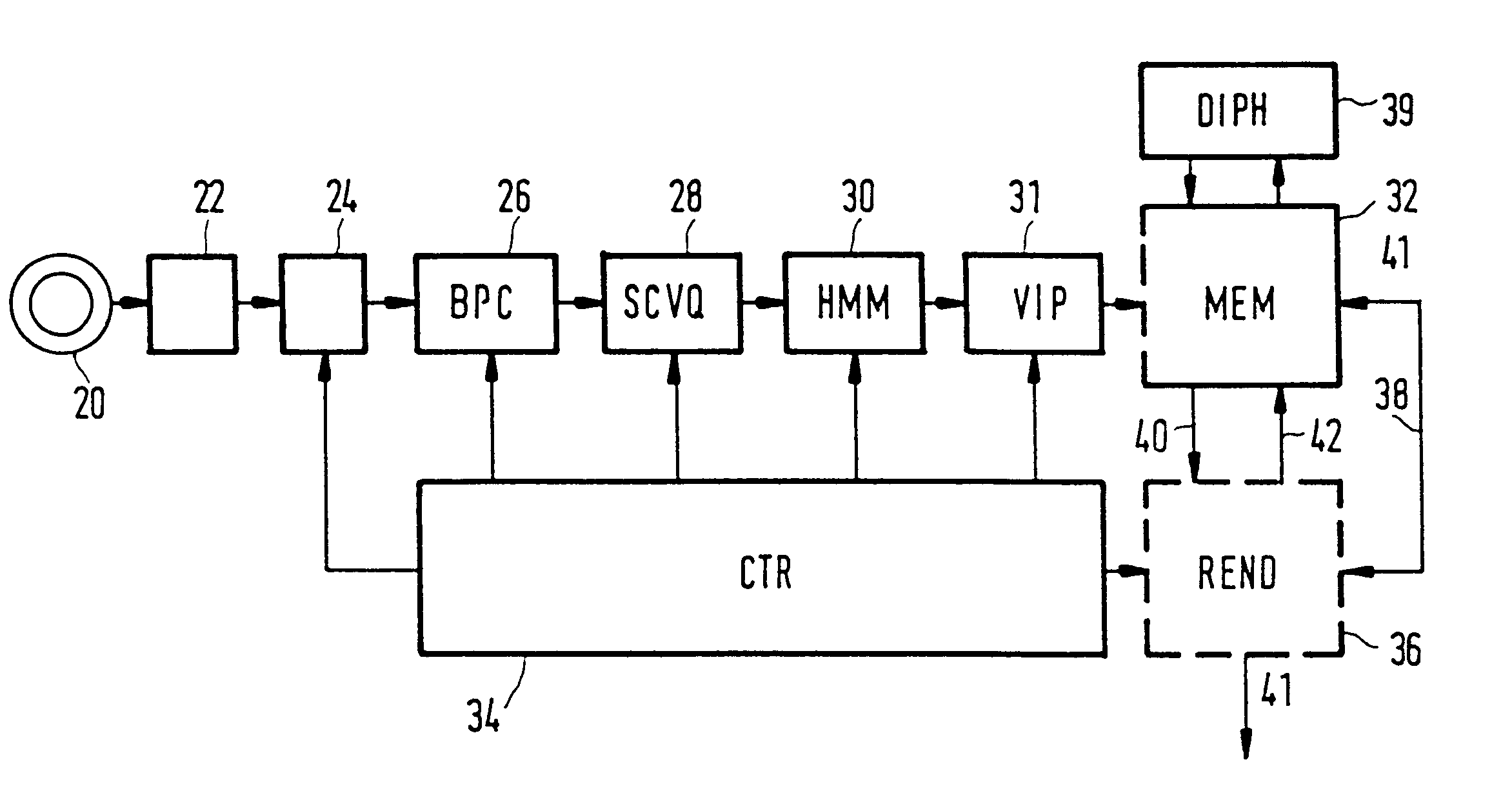
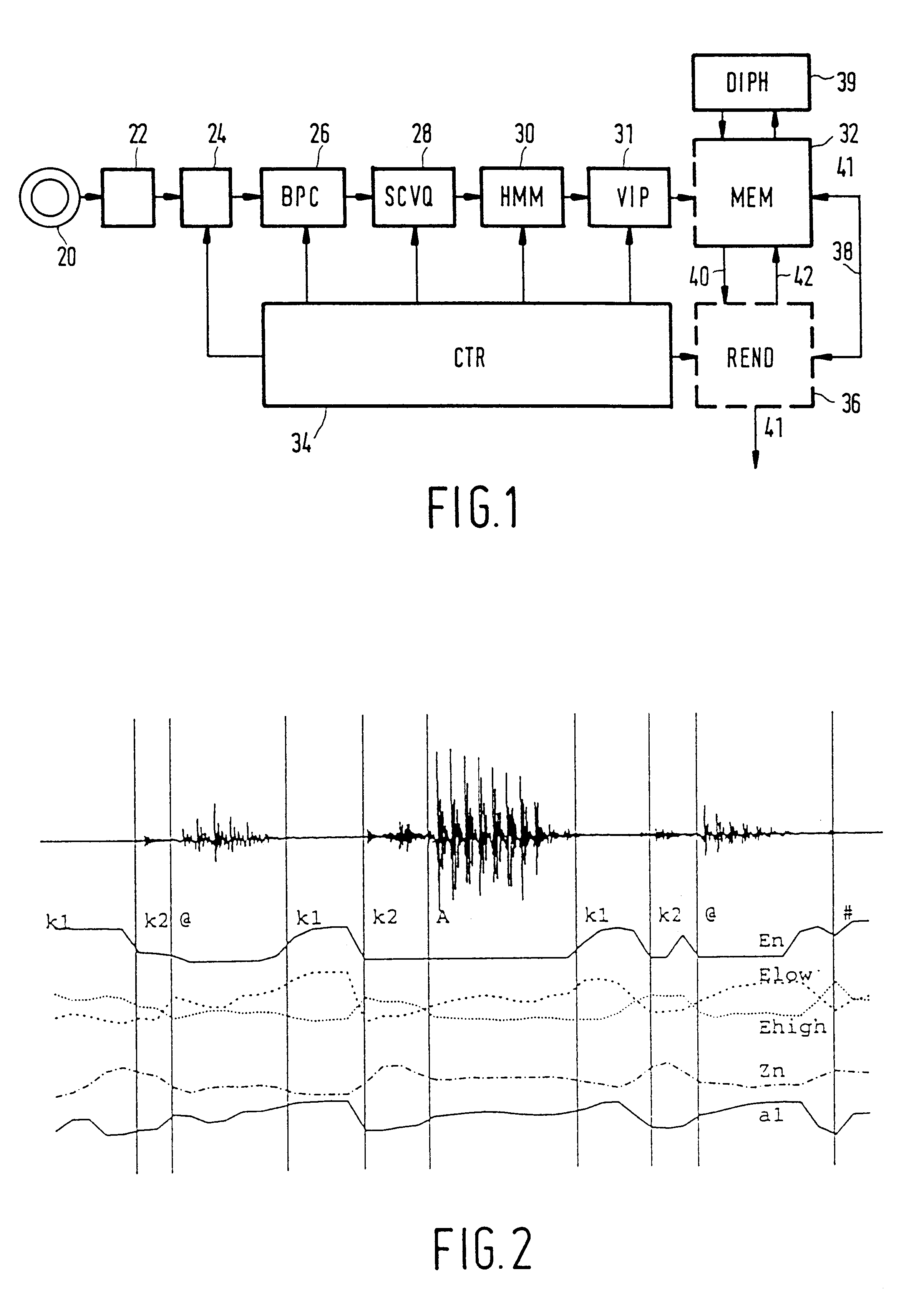
Method and apparatus for automatic speech segmentation into phoneme-like units for use in speech processing applications, and based on segmentation into broad phonetic classes, sequence-constrained vector quantization and hidden-markov-models
InactiveUS6208967B1Straightforward and inexpensiveDigital computer detailsBiological modelsAutomatic speech segmentationSpoken language
For machine segmenting of speech, first utterances from a database of known spoken words are classified and segmented into three broad phonetic classes (BPC) voiced, unvoiced, and silence. Next, using preliminary segmentation positions as anchor points, sequence-constrained vector quantization is used for further segmentation into phoneme-like units. Finally, exact tuning to the segmented phonemes is done through Hidden-Markov Modelling and after training a diphone set is composed for further usage.
Owner:U S PHILIPS CORP
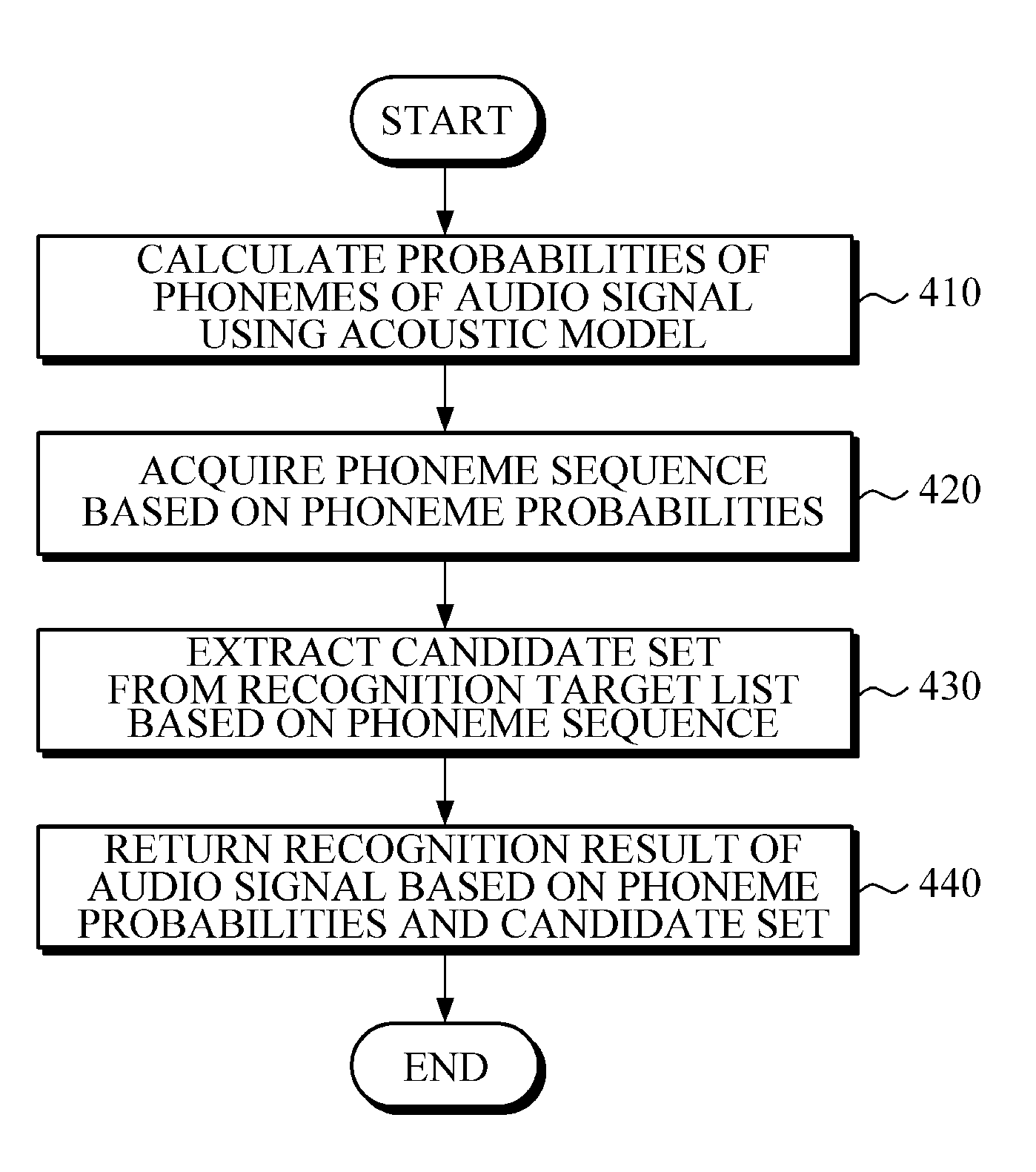
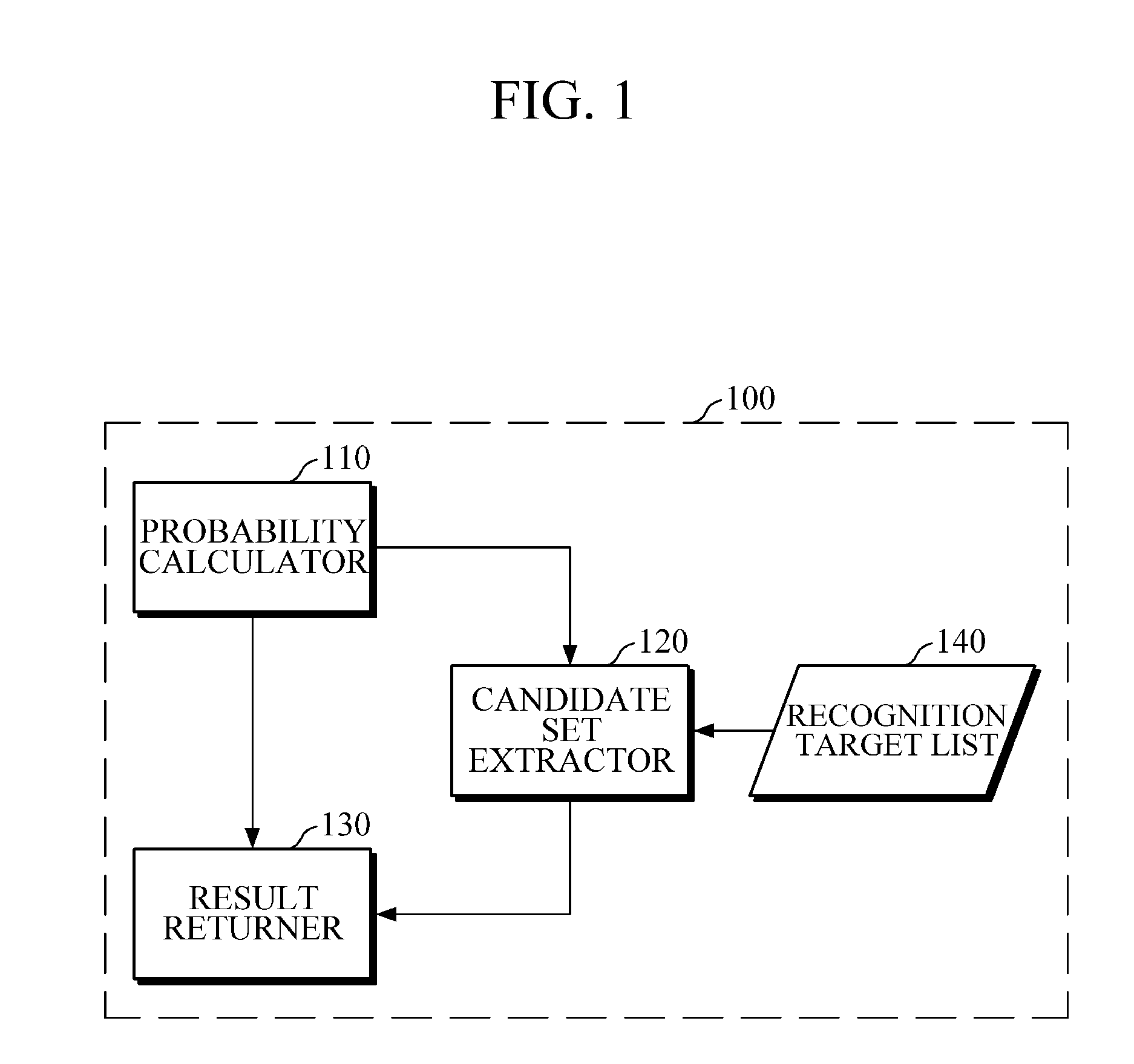
Speech recognition apparatus, speech recognition method, and electronic device
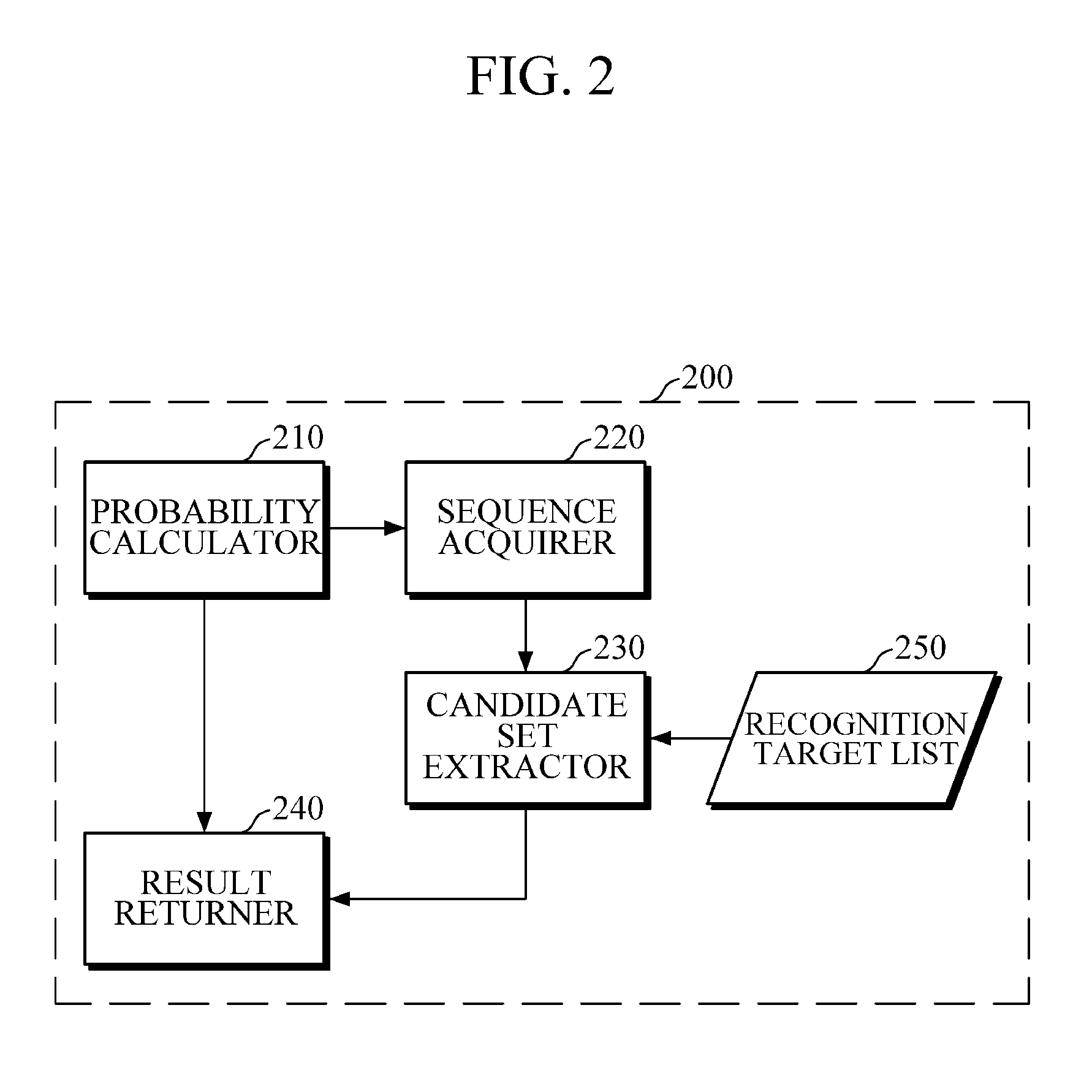
A speech recognition apparatus includes a probability calculator configured to calculate phoneme probabilities of an audio signal using an acoustic model; a candidate set extractor configured to extract a candidate set from a recognition target list; and a result returner configured to return a recognition result of the audio signal based on the calculated phoneme probabilities and the extracted candidate set.
Owner:SAMSUNG ELECTRONICS CO LTD
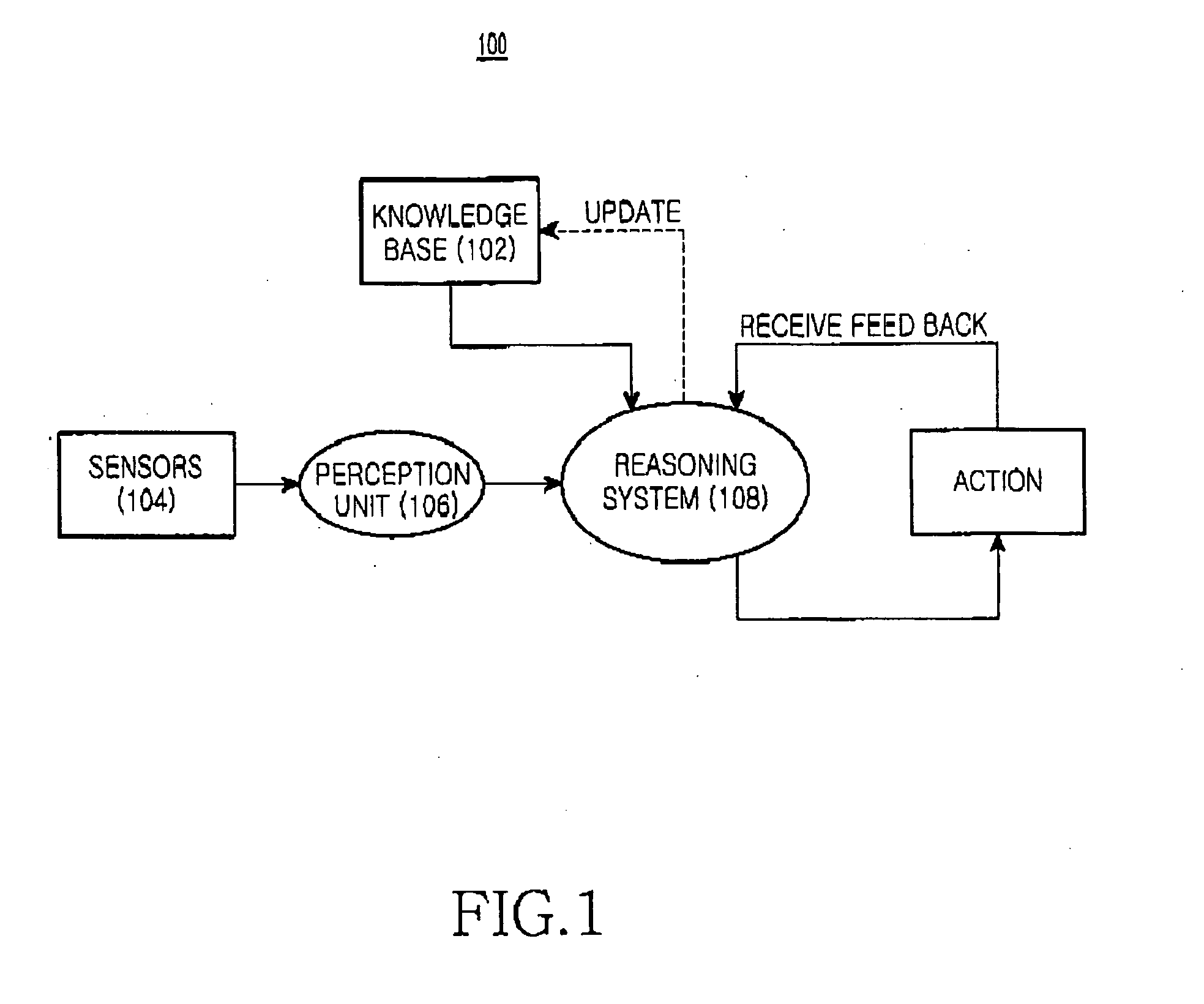
System and methods for adaptive control of robotic devices
ActiveUS7117067B2Improve efficiencyIncrease the number ofProgramme-controlled manipulatorDigital data processing detailsCommand and controlControl engineering
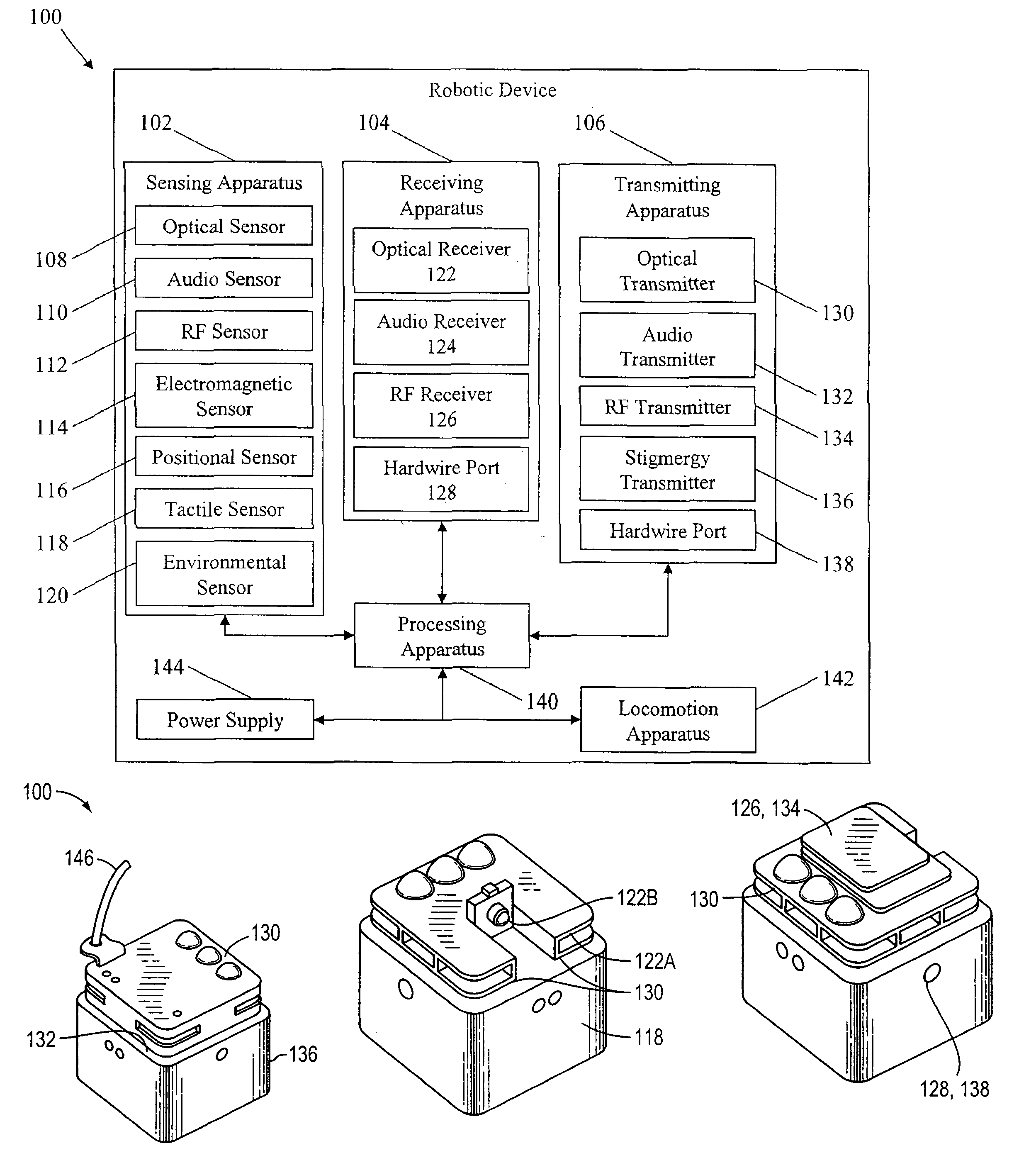
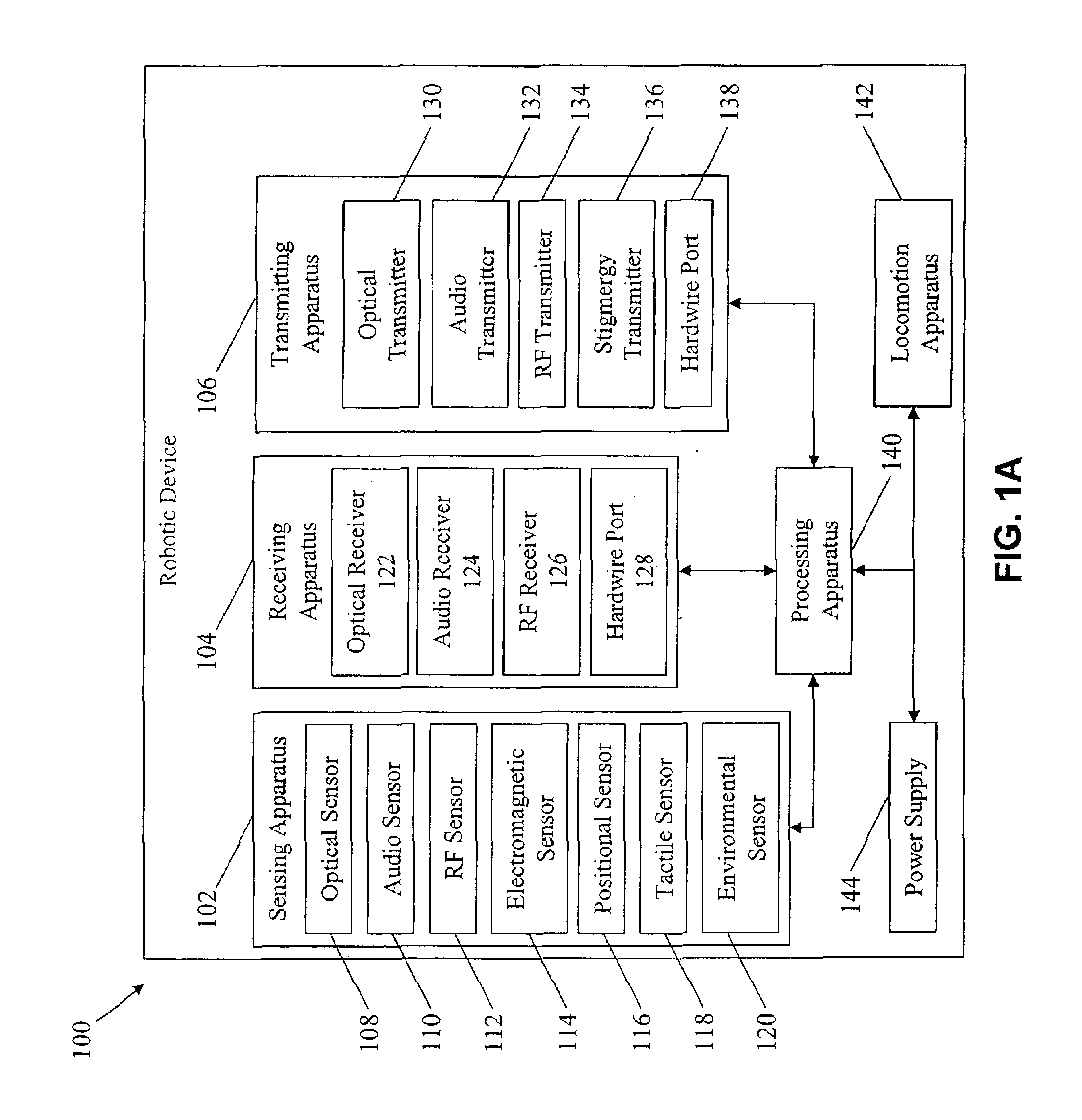
Methods for operating robotic devices (i.e., “robots”) that employ adaptive behavior relative to neighboring robots and external (e.g., environmental) conditions. Each robot is capable of receiving, processing, and acting on one or more multi-device primitive commands that describe a task the robot will perform in response to other robots and the external conditions. The commands facilitate a distributed command and control structure, relieving a central apparatus or operator from the need to monitor the progress of each robot. This virtually eliminates the corresponding constraint on the maximum number of robots that can be deployed to perform a task (e.g., data collection, mapping, searching). By increasing the number of robots, the efficiency in completing the task is also increased.
Owner:FLIR DETECTION
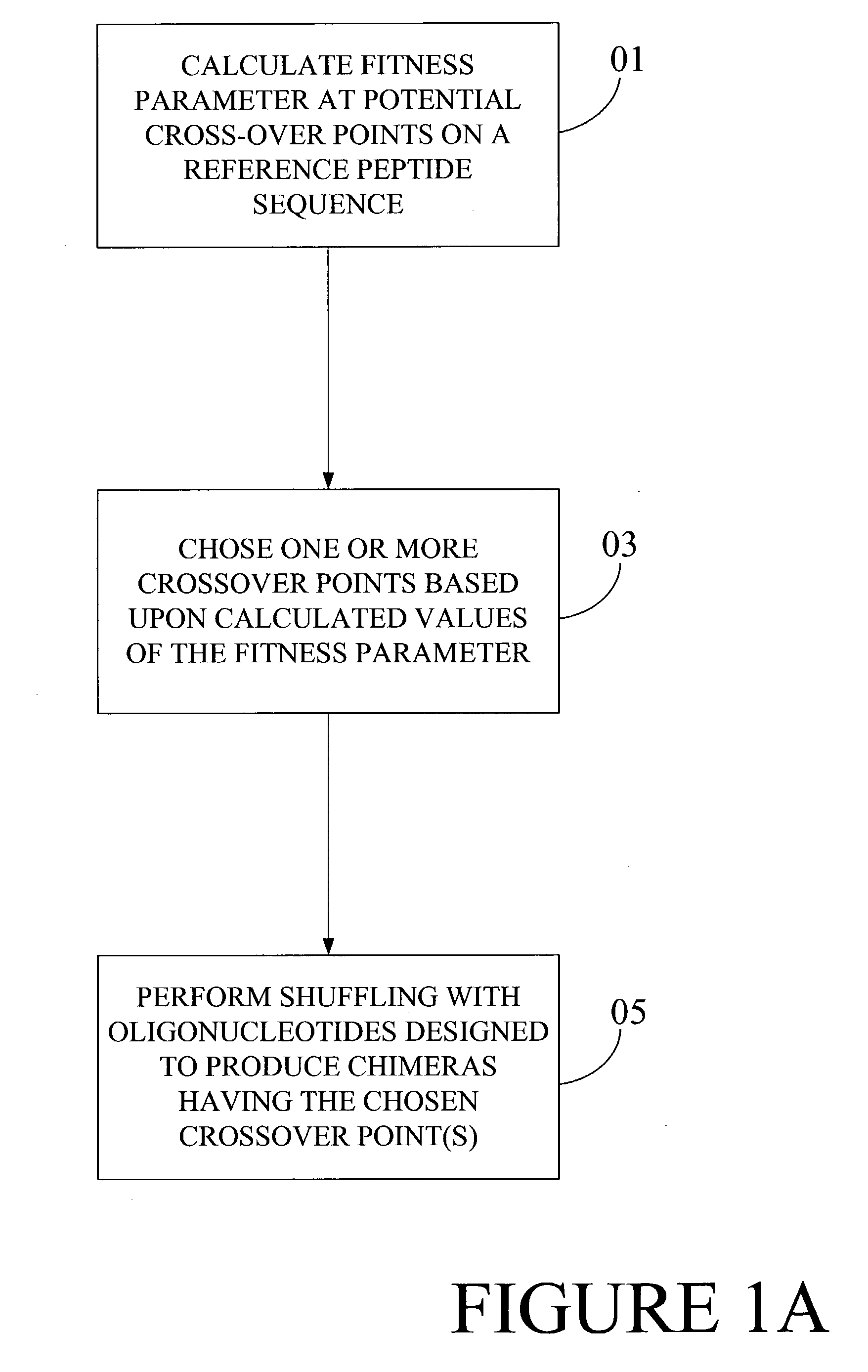
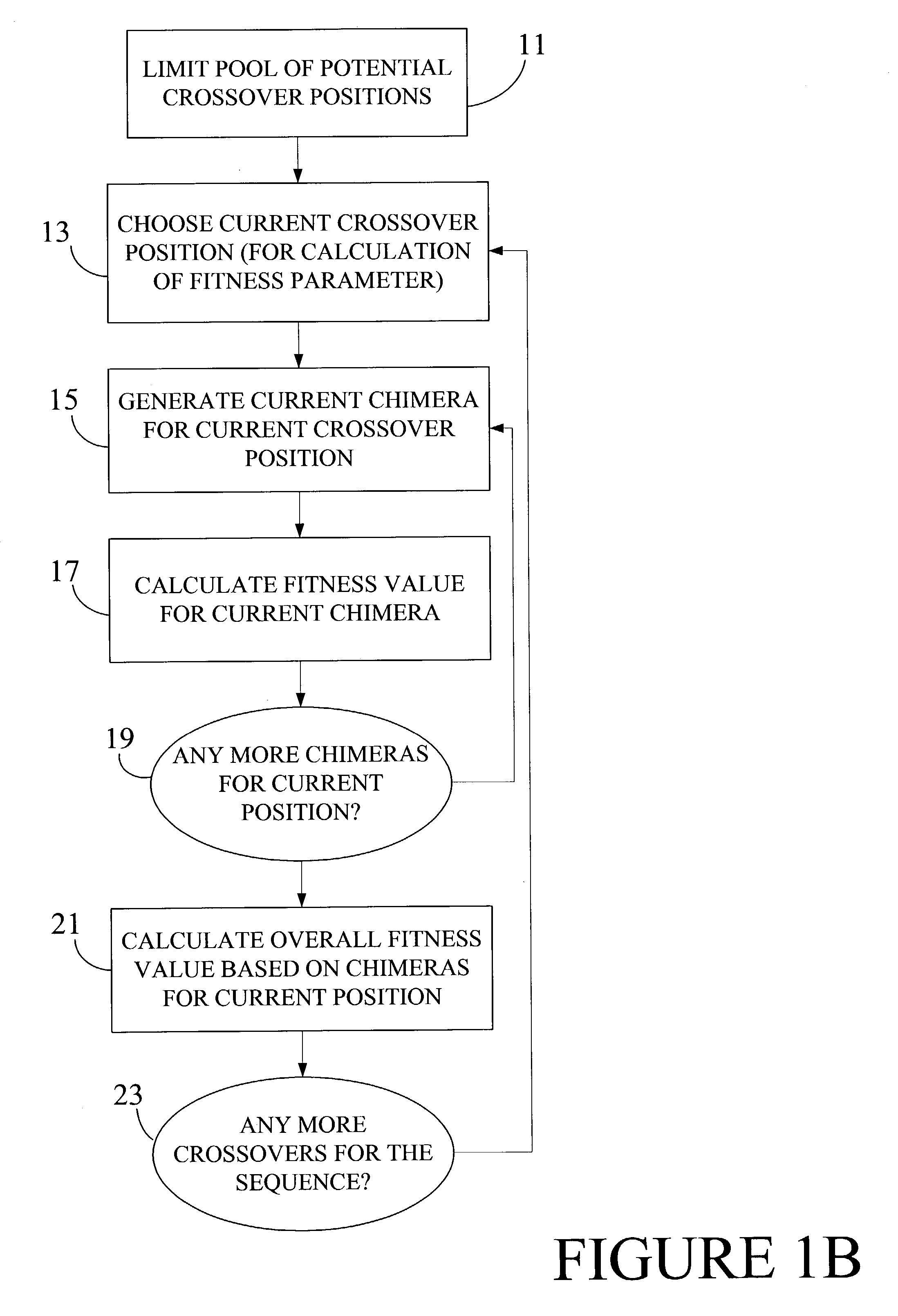
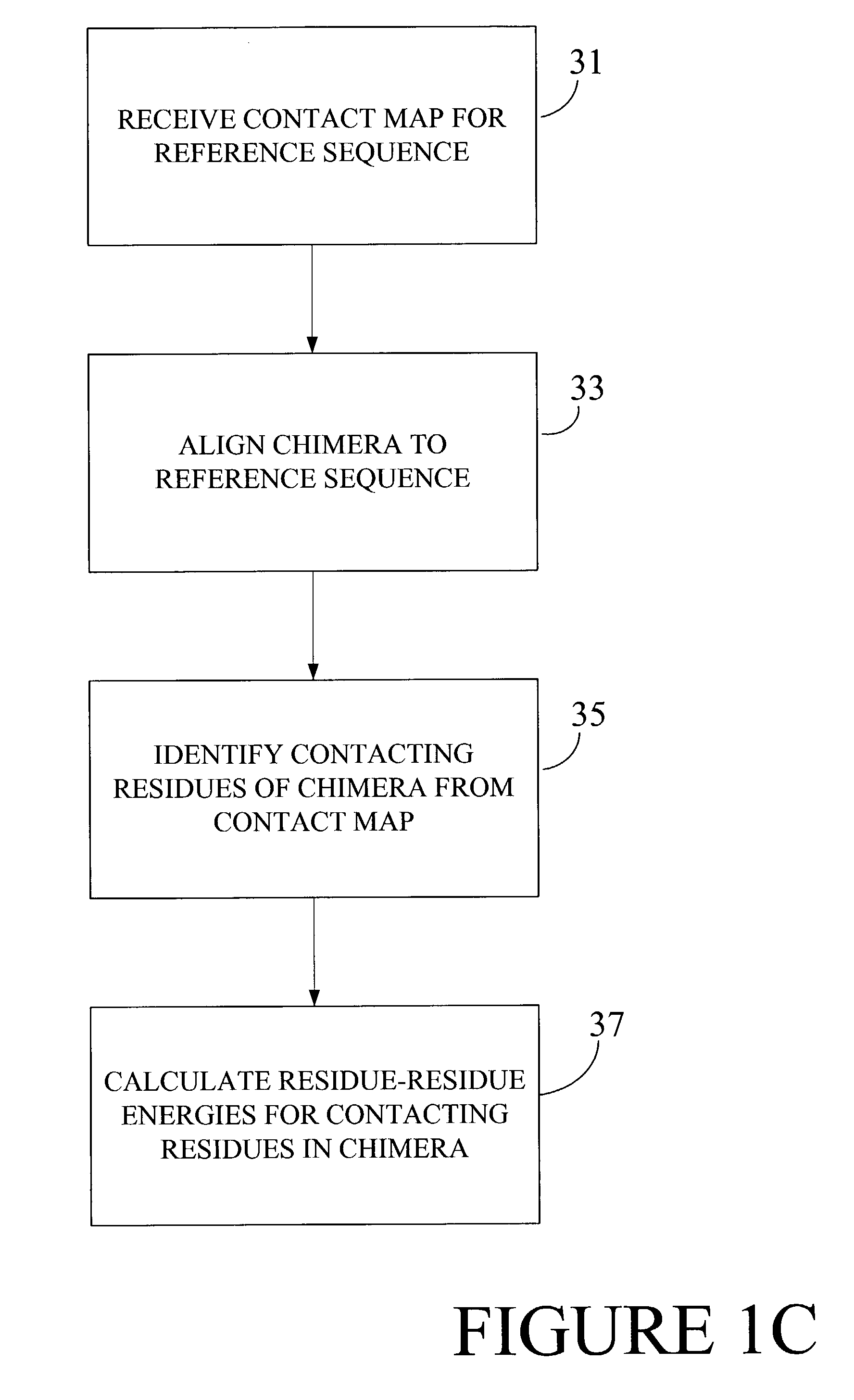
Optimization of crossover points for directed evolution
ActiveUS7620500B2Microbiological testing/measurementAnalogue computers for chemical processesNucleotideNucleotide sequencing
Owner:CODEXIS MAYFLOWER HLDG LLC
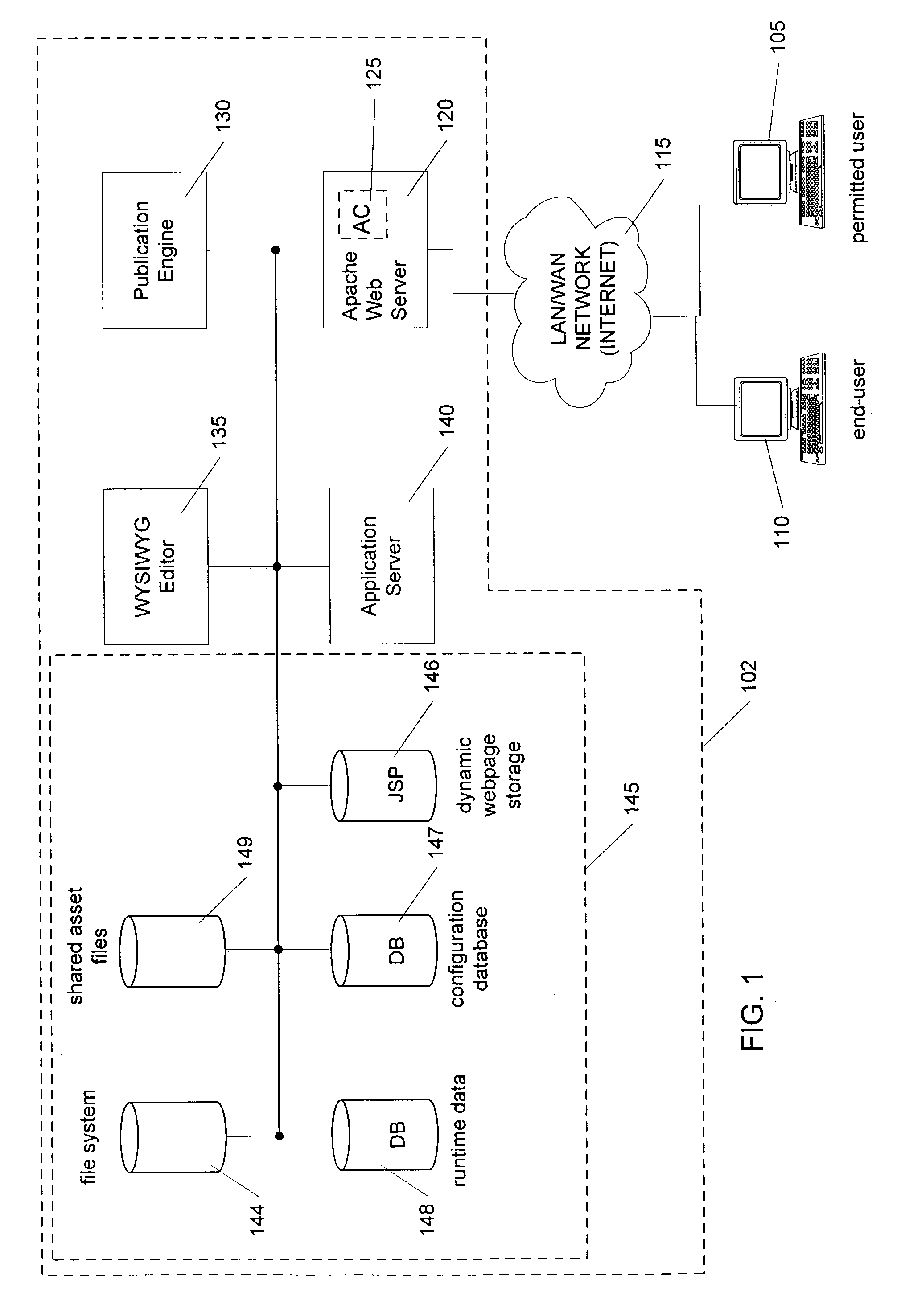
Method and apparatus for processing a dynamic webpage
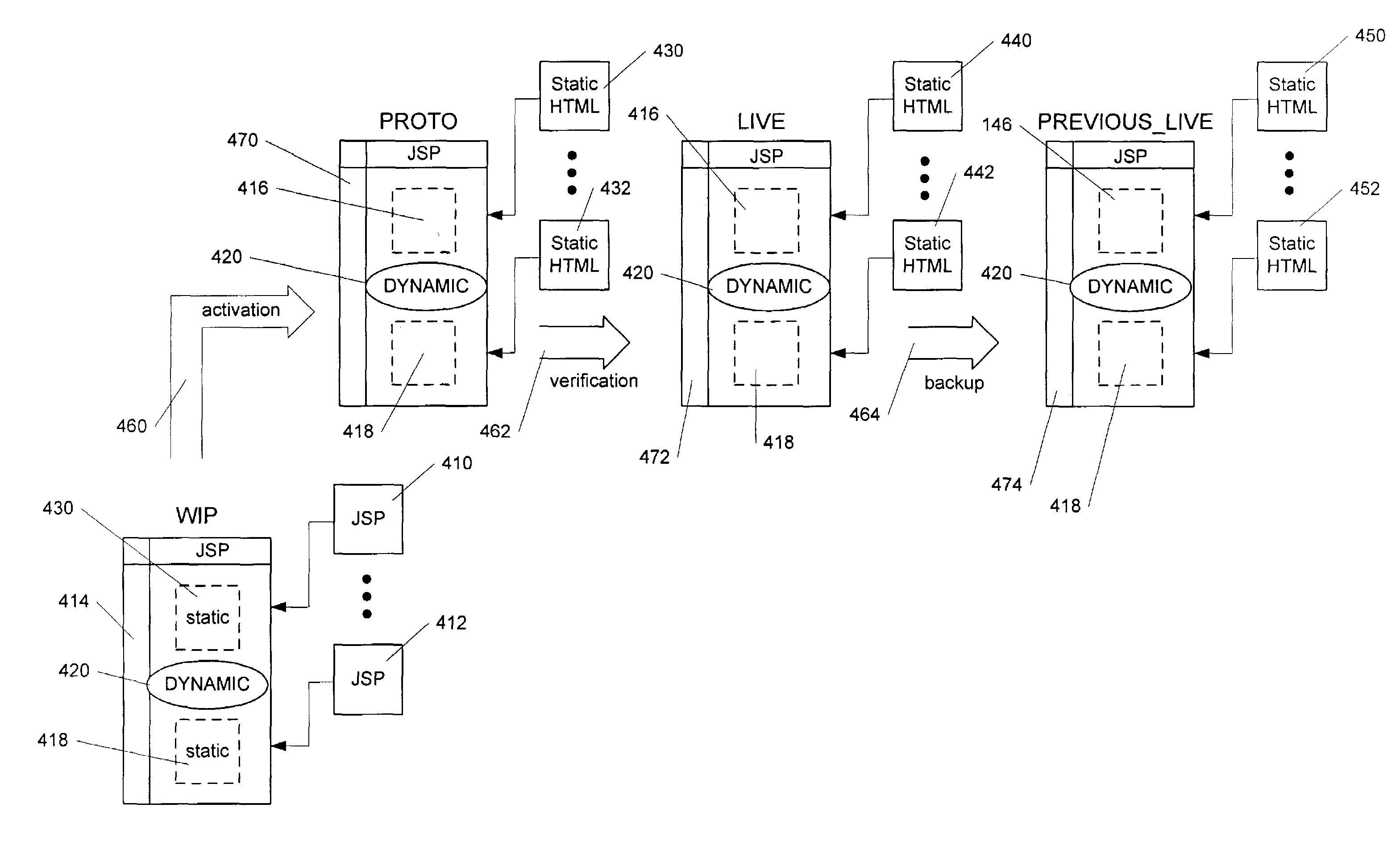
InactiveUS7386786B2Reduce processing requirementsDigital computer detailsBiological modelsProgramming languageDynamic web page
A dynamic webpage processing method, including identifying a static component within a dynamic webpage, the static component comprising computer code defining at least a portion of a target webpage and configurable via first configuration data, rendering the static component as a static markup language equivalent, and returning the target webpage upon request through rendering a dynamic component within the dynamic webpage based on second configuration data is disclosed. Thus, static component(s) can be pre-rendered, leaving only the dynamic component(s) of the dynamic webpage for on-the-fly rendering when a target webpage request is made. This can greatly reduce dynamic webpage processing requirements, particularly in situations where the webpage configuration changes or evolves slowly.
Owner:SINCRO LLC
Method and apparatus for synchronizing multiple versions of digital data
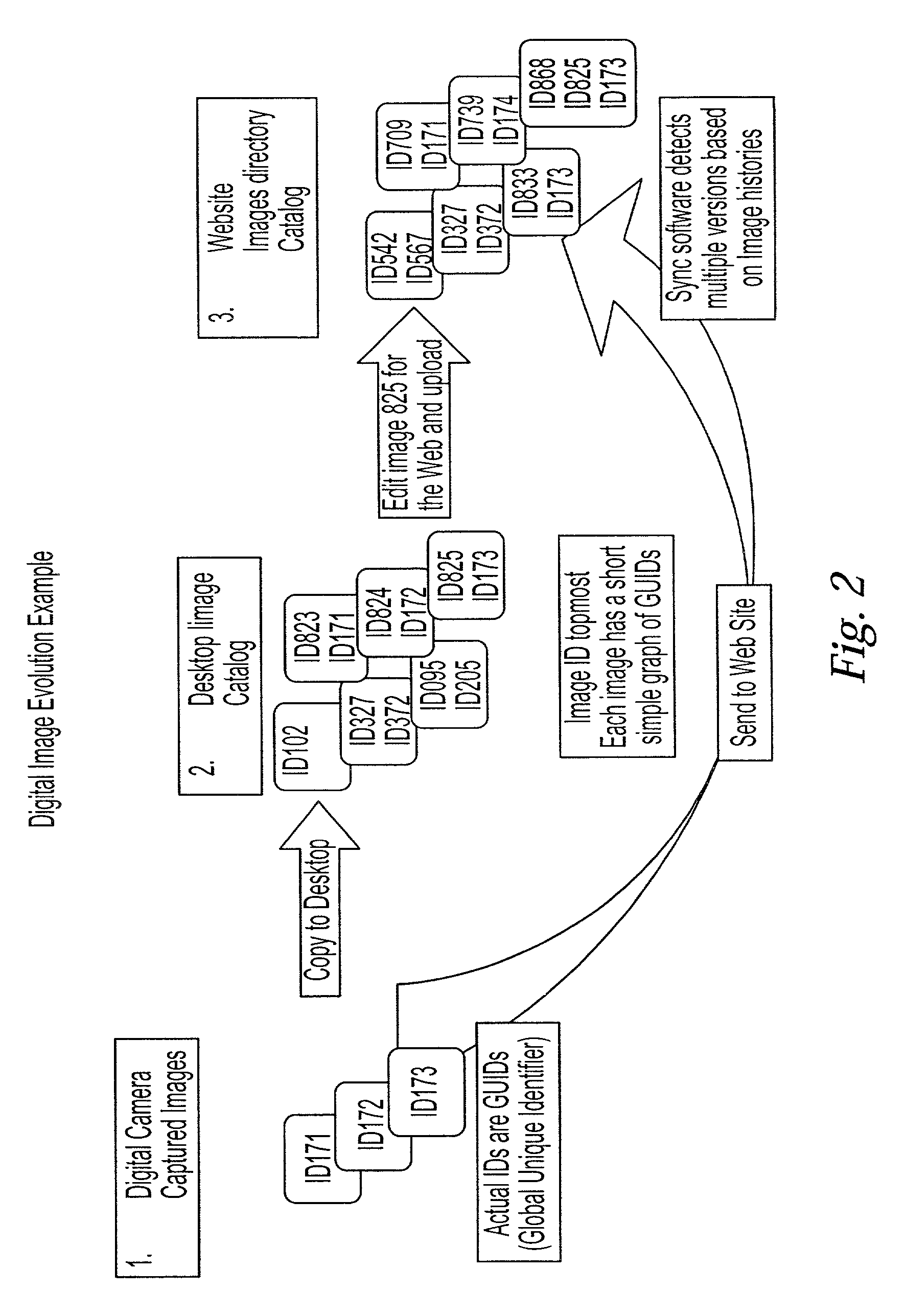
InactiveUS7216289B2Reduce overheadGuaranteed to workData processing applicationsDigital data information retrievalDigital dataUnique identifier
A method and system for synchronizing multiple versions of multimedia objects is provided. Each multimedia object may be identified by a unique identifier. In addition, a history graph may be generated and maintained for each object, where the history graph includes nodes that store unique identifiers and whose vectors describe the relationship between the multimedia objects. Metadata may be used to describe the transformations of objects.
Owner:MICROSOFT TECH LICENSING LLC
A session robotic system
InactiveCN101187990ARecognizableHave the ability to understandInput/output for user-computer interactionBiological modelsRobotic systemsSpeech identification
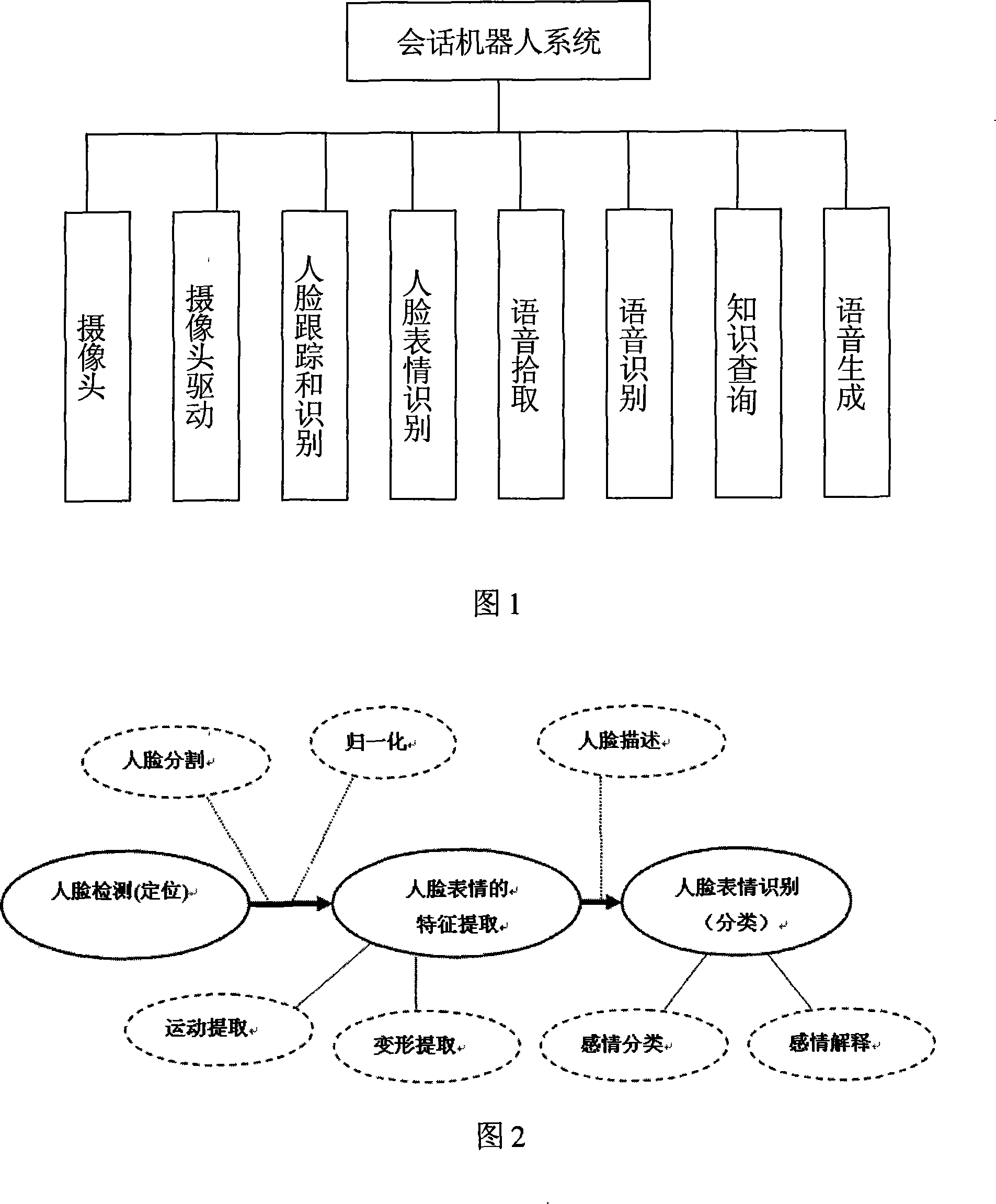
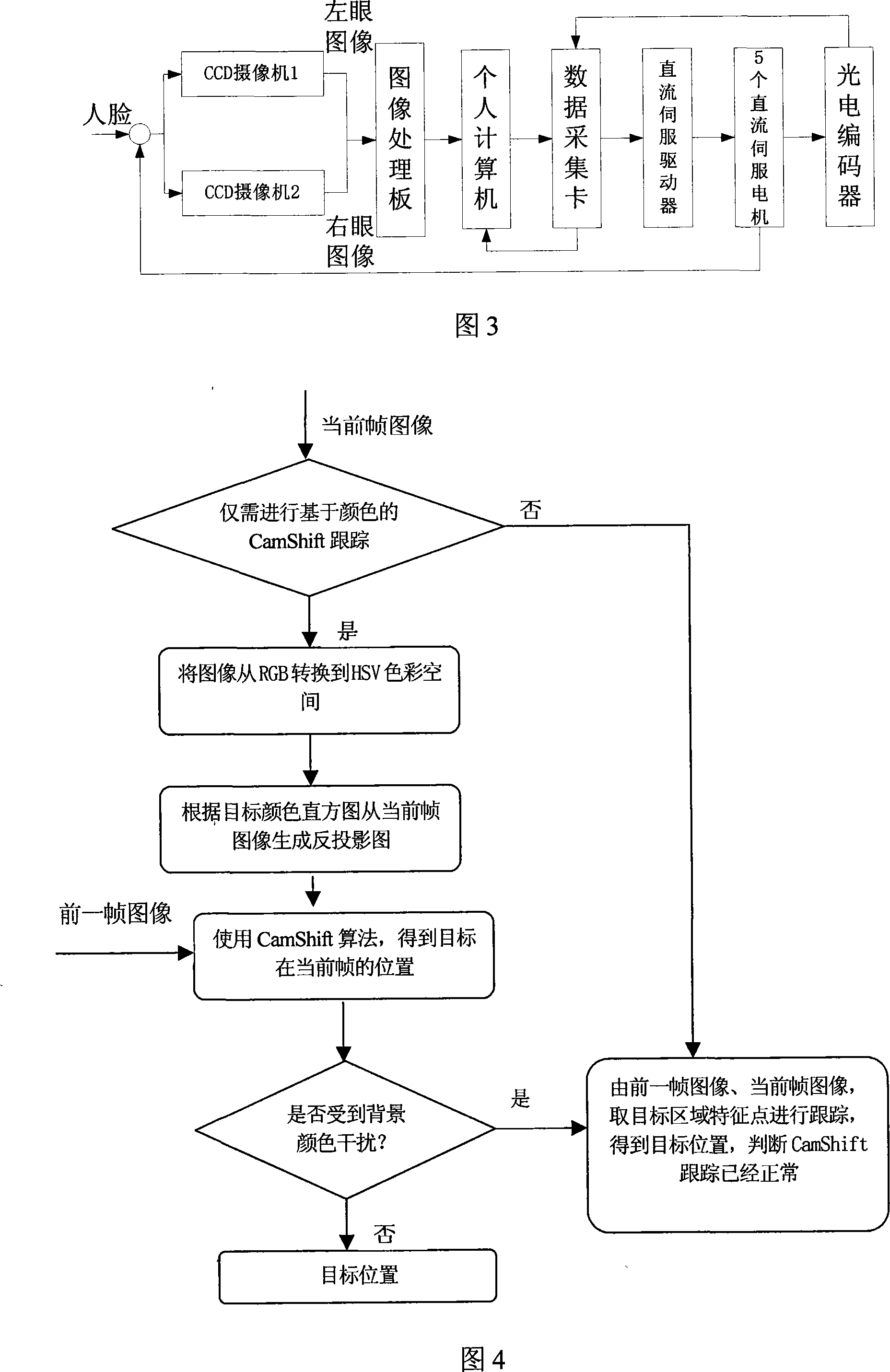
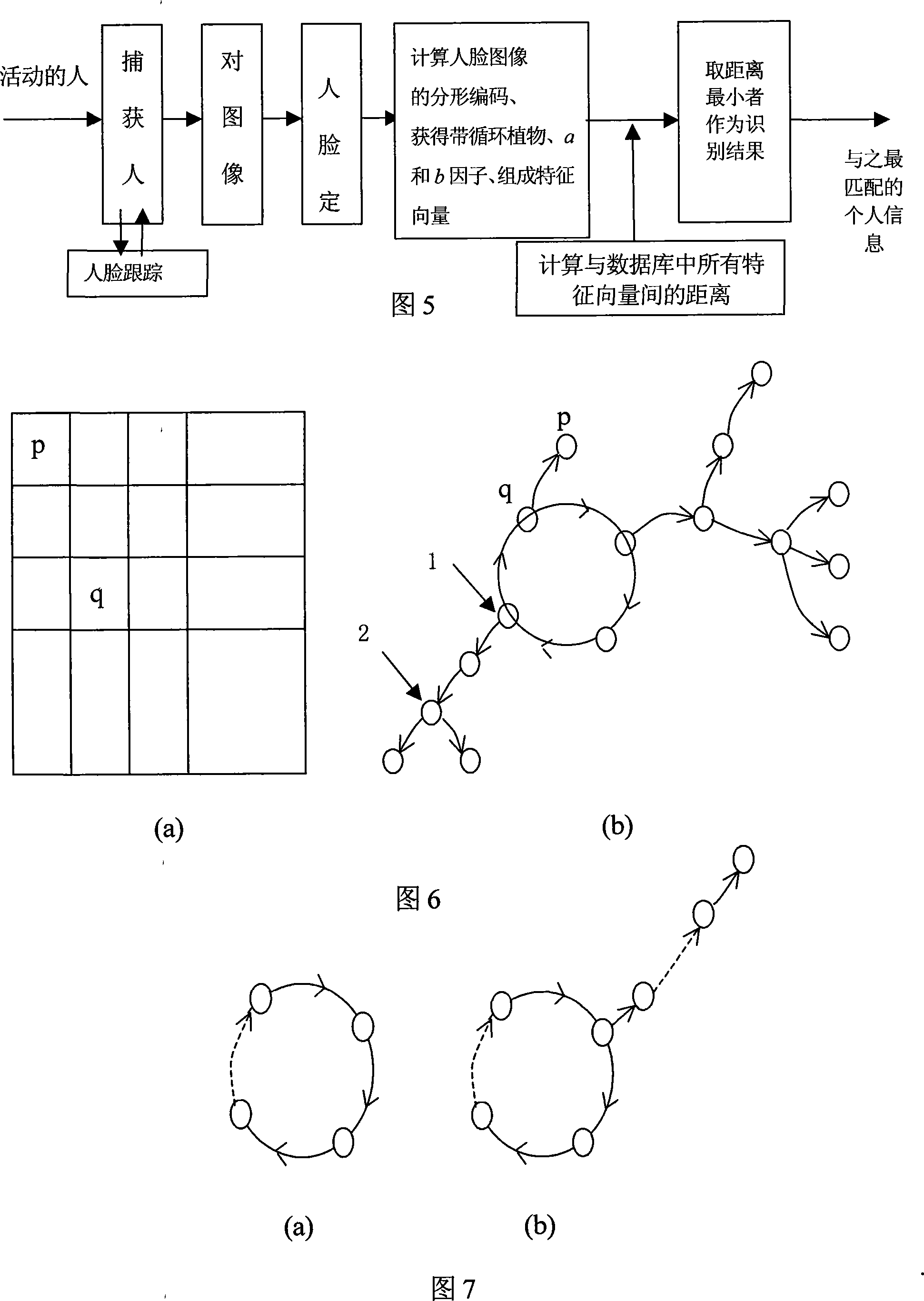
The invention discloses a conservational robot system. A human face tracking and recognizing module tracks and recognizes a human face image captured by a camera; a human facial expression recognizing module recognizes the expression; and semantic meaning is recognized after voice signals pass through a voice picking module and a voice recognizing module. The robot system understands human demands according to facial expressions and / or voice and then forms conservation statement through a knowledge inquiry module and generates voice for communication with humans through a voice generating module. The conservational robot system has voice recognizing and understanding abilities and can understand commands of users. The invention can be applied to schools, families, hotels, companies, airports, bus stations, docks, meeting rooms and so on for education, chat, conservation, consultation, etc. In addition, the invention can also help users with propaganda and introduction, guest reception, business inquiry, secretary service, foreign language interpretation, etc.
Owner:SOUTH CHINA UNIV OF TECH
Systems and methods for quantitative analysis of histopathology images using multiclassifier ensemble schemes
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV +1
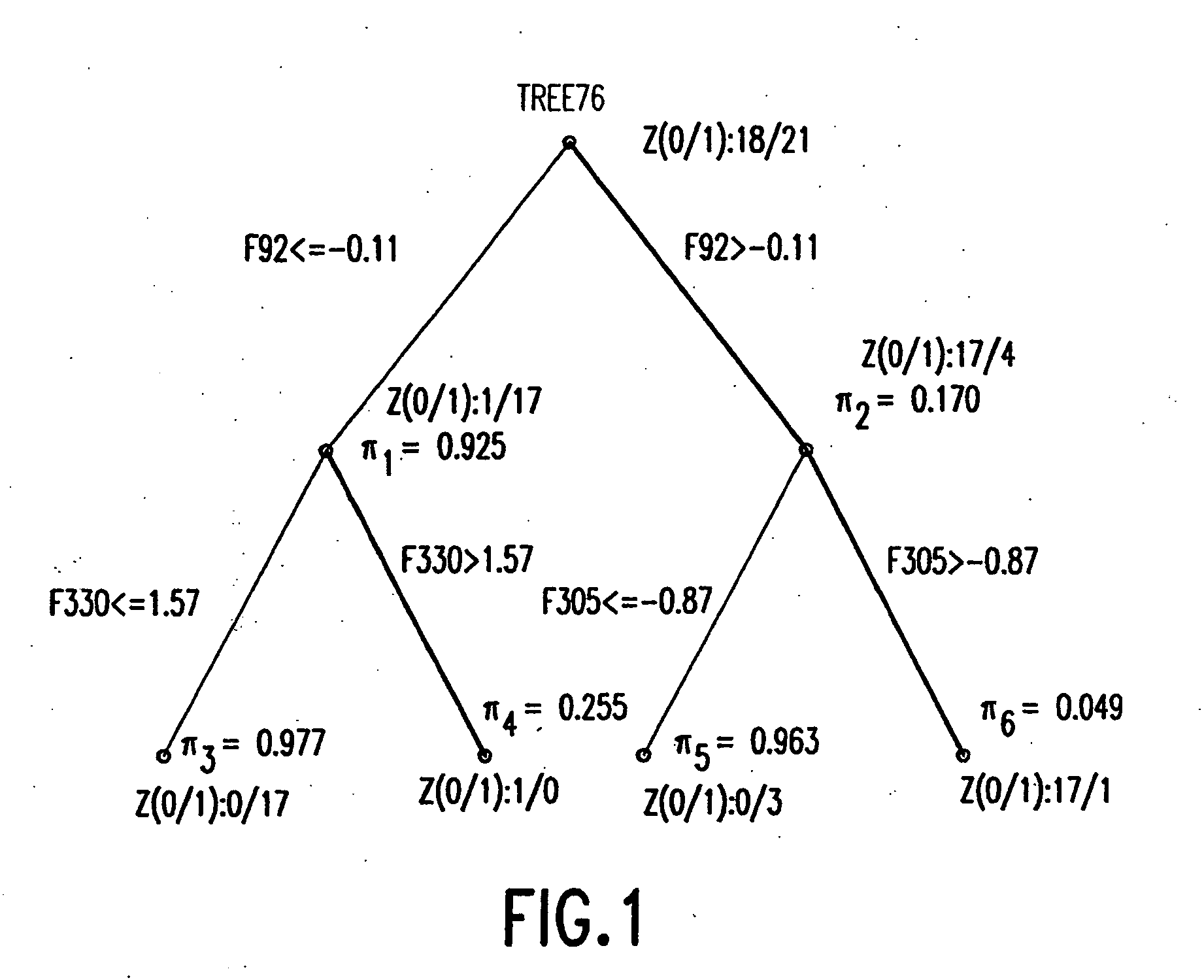
Binary prediction tree modeling with many predictors and its uses in clinical and genomic applications
The statistical analysis described and claimed is a predictive statistical tree model that overcomes several problems observed in prior statistical models and regression analyses, while ensuring greater accuracy and predictive capabilities. Although the claimed use of the predictive statistical tree model described herein is directed to the prediction of a disease in individuals, the claimed model can be used for a variety of applications including the prediction of disease states, susceptibility of disease states or any other biological state of interest, as well as other applicable non-biological states of interest. This model first screens genes to reduce noise, applies k-means correlation-based clustering targeting a large number of clusters, and then uses singular value decompositions (SVD) to extract the single dominant factor (principal component) from each cluster. This generates a statistically significant number of cluster-derived singular factors, that we refer to as metagenes, that characterize multiple patterns of expression of the genes across samples. The strategy aims to extract multiple such patterns while reducing dimension and smoothing out gene-specific noise through the aggregation within clusters. Formal predictive analysis then uses these metagenes in a Bayesian classification tree analysis. This generates multiple recursive partitions of the sample into subgroups (the “leaves” of the classification tree), and associates Bayesian predictive probabilities of outcomes with each subgroup. Overall predictions for an individual sample are then generated by averaging predictions, with appropriate weights, across many such tree models. The model includes the use of iterative out-of-sample, cross-validation predictions leaving each sample out of the data set one at a time, refitting the model from the remaining samples and using it to predict the hold-out case. This rigorously tests the predictive value of a model and mirrors the real-world prognostic context where prediction of new cases as they arise is the major goal.
Owner:DUKE UNIV
Extensible stylesheet designs using meta-tag and/or associated meta-tag information
Techniques for extensible stylesheet designs using meta-tag and / or associated meta-tag Information are described. To generate a proper stylesheet (e.g., an XSL or XSLT file) from a source file (e.g., an XML file), all meta-tag and / or associated meta-tag Information are differentiated by attaching respectively unique identifiers to those that are otherwise identical. To facilitate user required operations on certain data in the source file, a document source path for the data is identified and inserted with one or more operators thus to form document source path information. The differentiated meta-tag and / or associated meta-tag Information and source path information are relied upon to generate one or more stylesheets.
Owner:CHARTOLEAUX
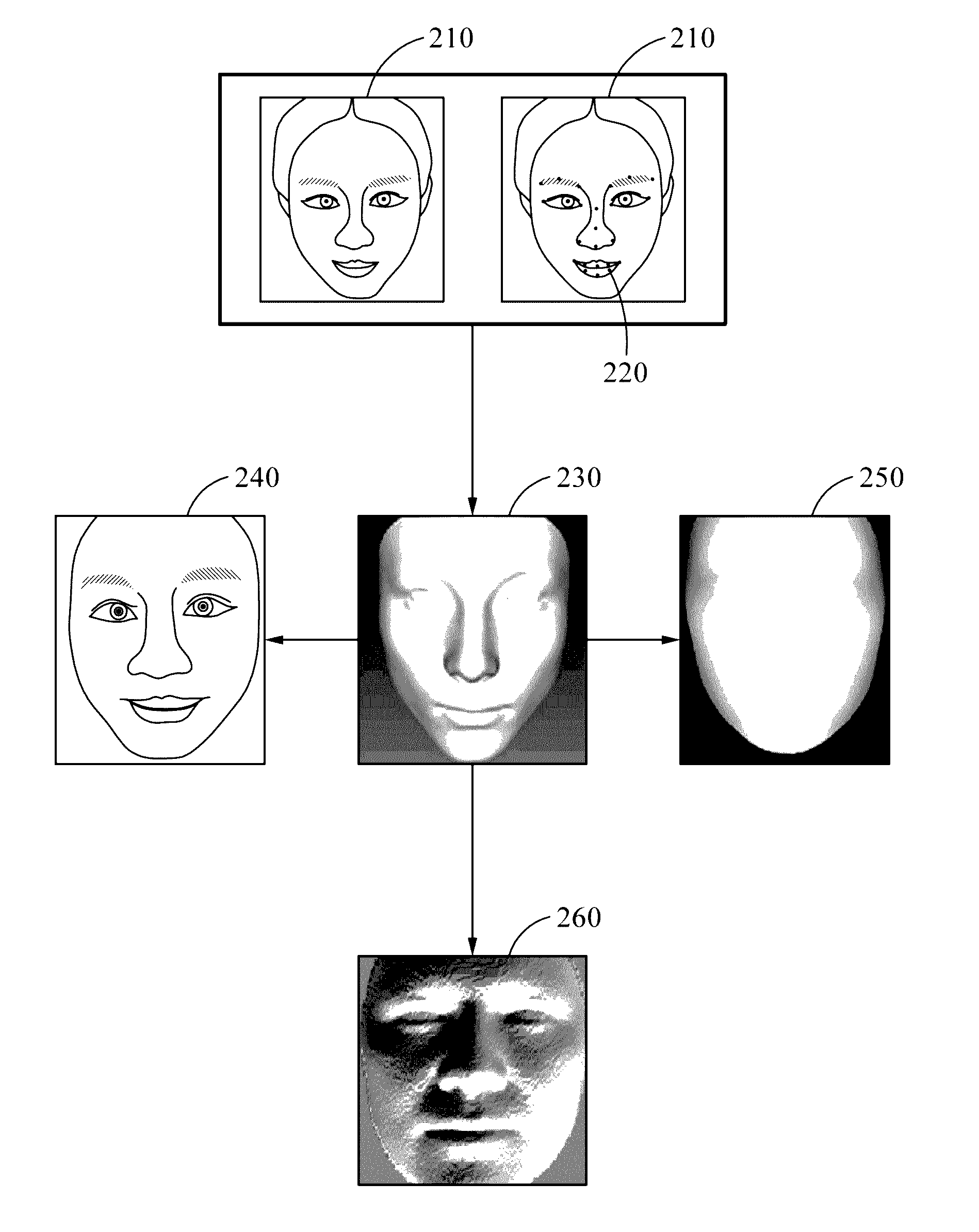
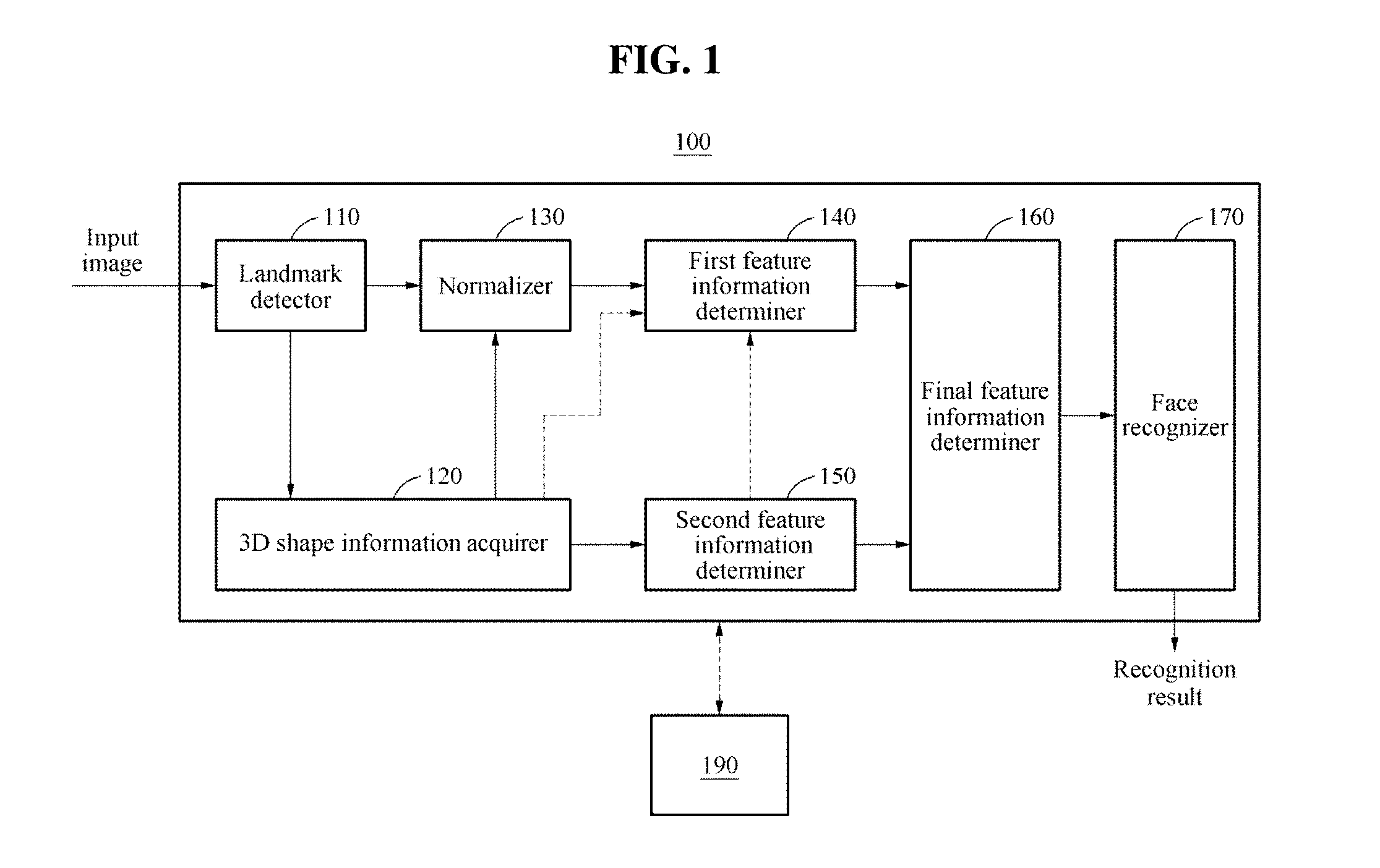
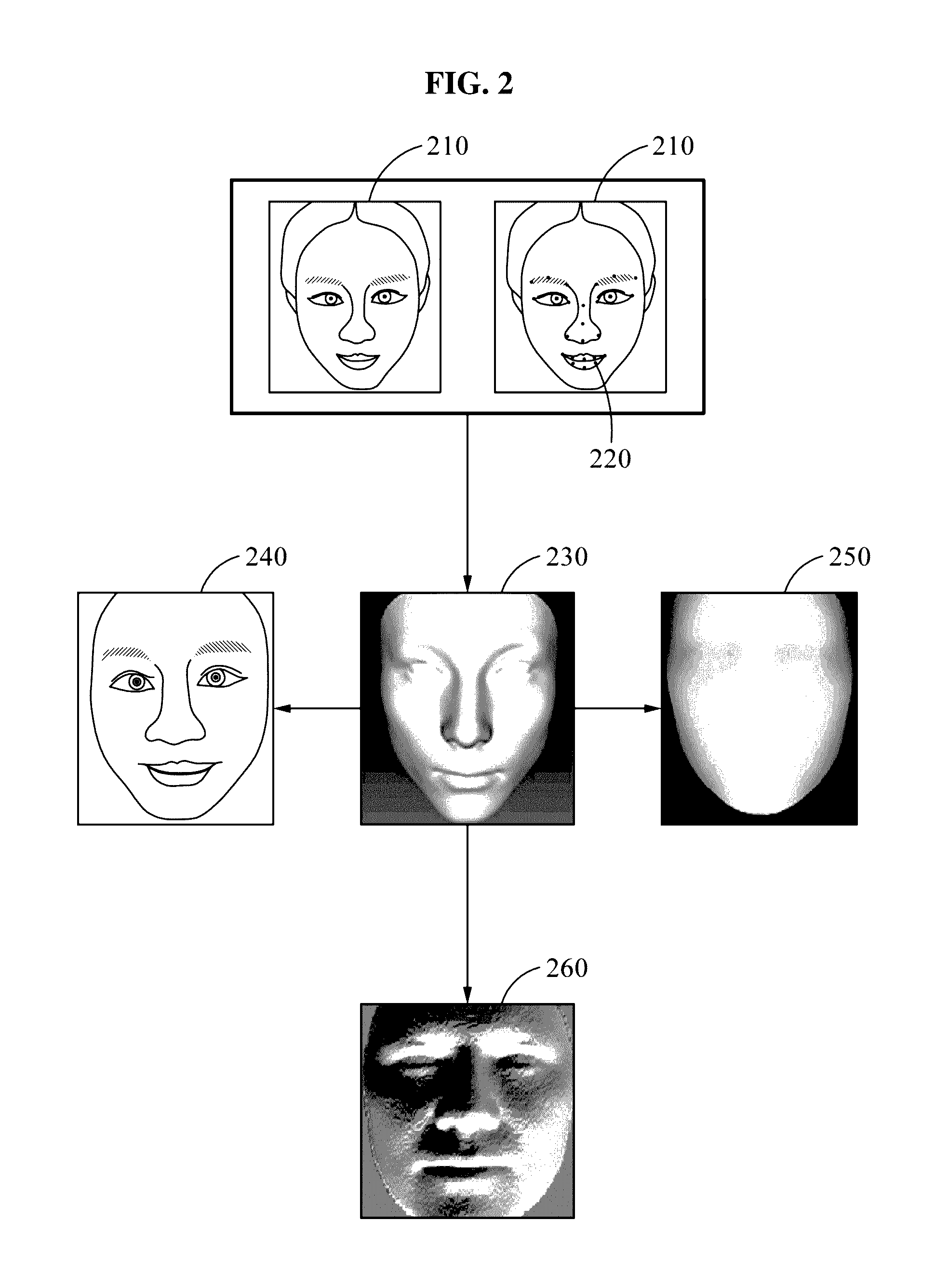
Face recognition method and apparatus
Face recognition of a face, to determine whether the face correlates with an enrolled face, may include generating a personalized three-dimensional (3D) face model based on a two-dimensional (2D) input image of the face, acquiring 3D shape information and a normalized 2D input image of the face based on the personalized 3D face model, generating feature information based on the 3D shape information and pixel color values of the normalized 2D input image, and comparing the feature information with feature information associated with the enrolled face. The feature information may include first and second feature information generated based on applying first and second deep neural network models to the pixel color values of the normalized 2D input image and the 3D shape information, respectively. The personalized 3D face model may be generated based on transforming a generic 3D face model based on landmarks detected in the 2D input image.
Owner:SAMSUNG ELECTRONICS CO LTD
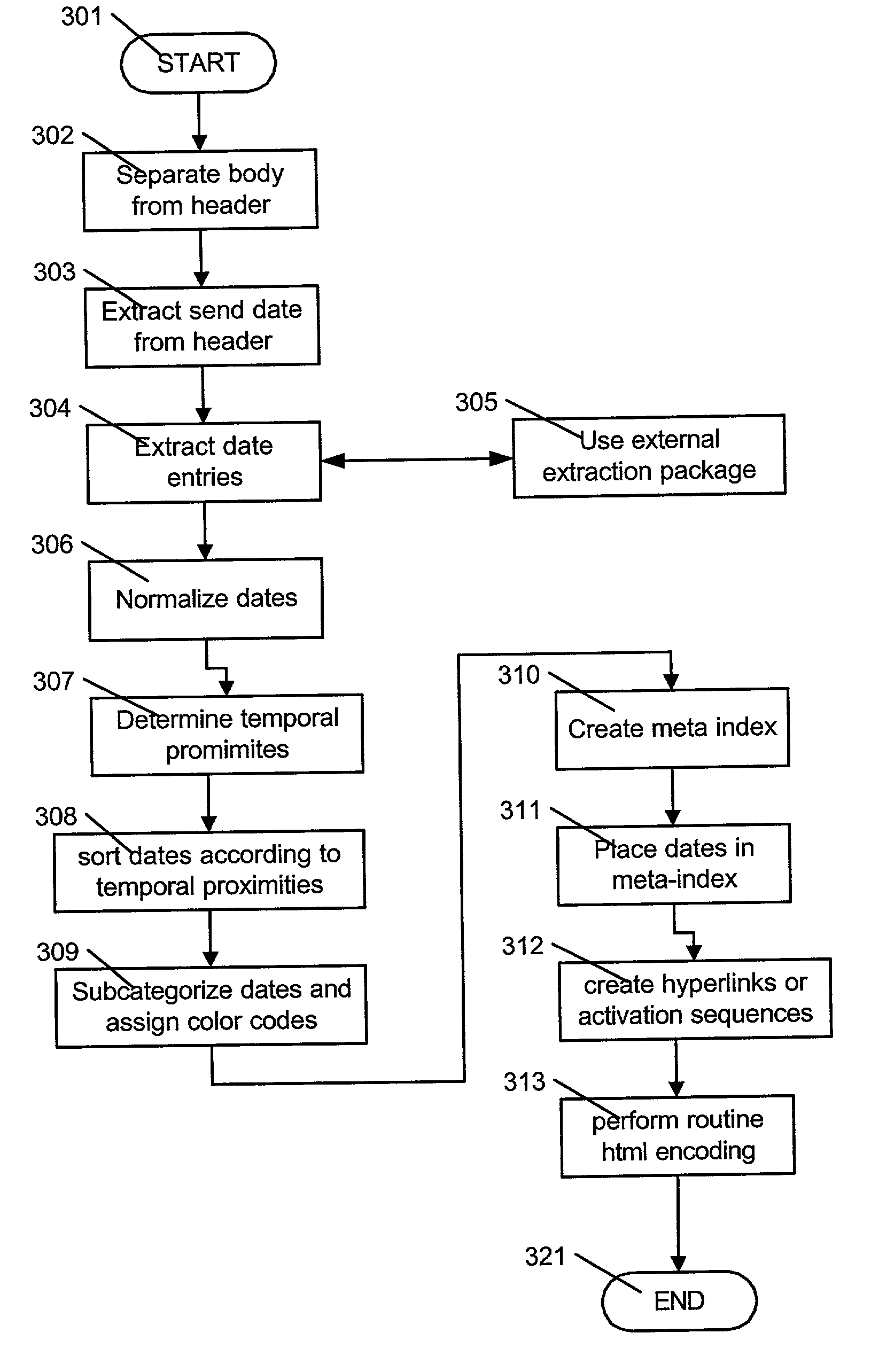
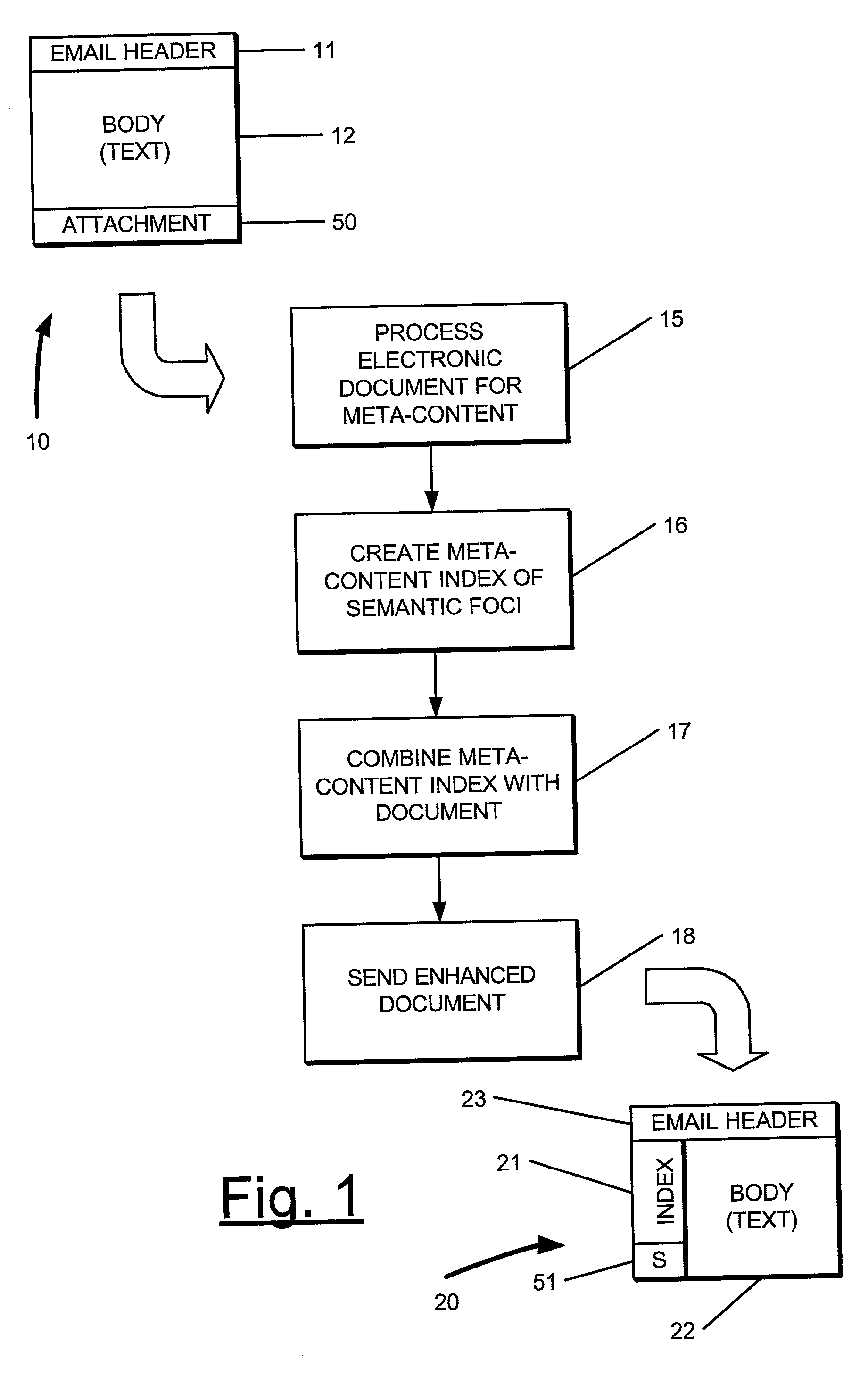
Meta-content analysis and annotation of email and other electronic documents
Meta-content analysis and annotation upon the body of email documents, and other electronic documents, and to create a displayable index of these instances of meta-content, which is sorted and annotated by type are provided. In addition, the electronic document is enhanced by providing links for the semantic foci to external documents containing related information. An electronic document adapted for delivery to one or more recipients, the electronic document including a header and a body, is processed by:performing meta-content extraction of semantic foci within said header and said body, the semantic foci comprising a plurality of type of information including one or more of email addresses, URLs, dates, currency values, organization names, names of people, names of places, and phone numbers;creating a meta-content index the document based upon said extracted semantic foci;arranging the meta-index according to said plurality of types;combining said meta-content index with said header and said body to provide an enhanced document; andsending said enhanced document to said one or more recipients via a communication network.The process includes converting the electronic mail document to a markup language format, and wherein said meta-content index comprises one or more objects expressed in said markup language adapted for presentation with body in said enhanced document.
Owner:SAP AMERICA
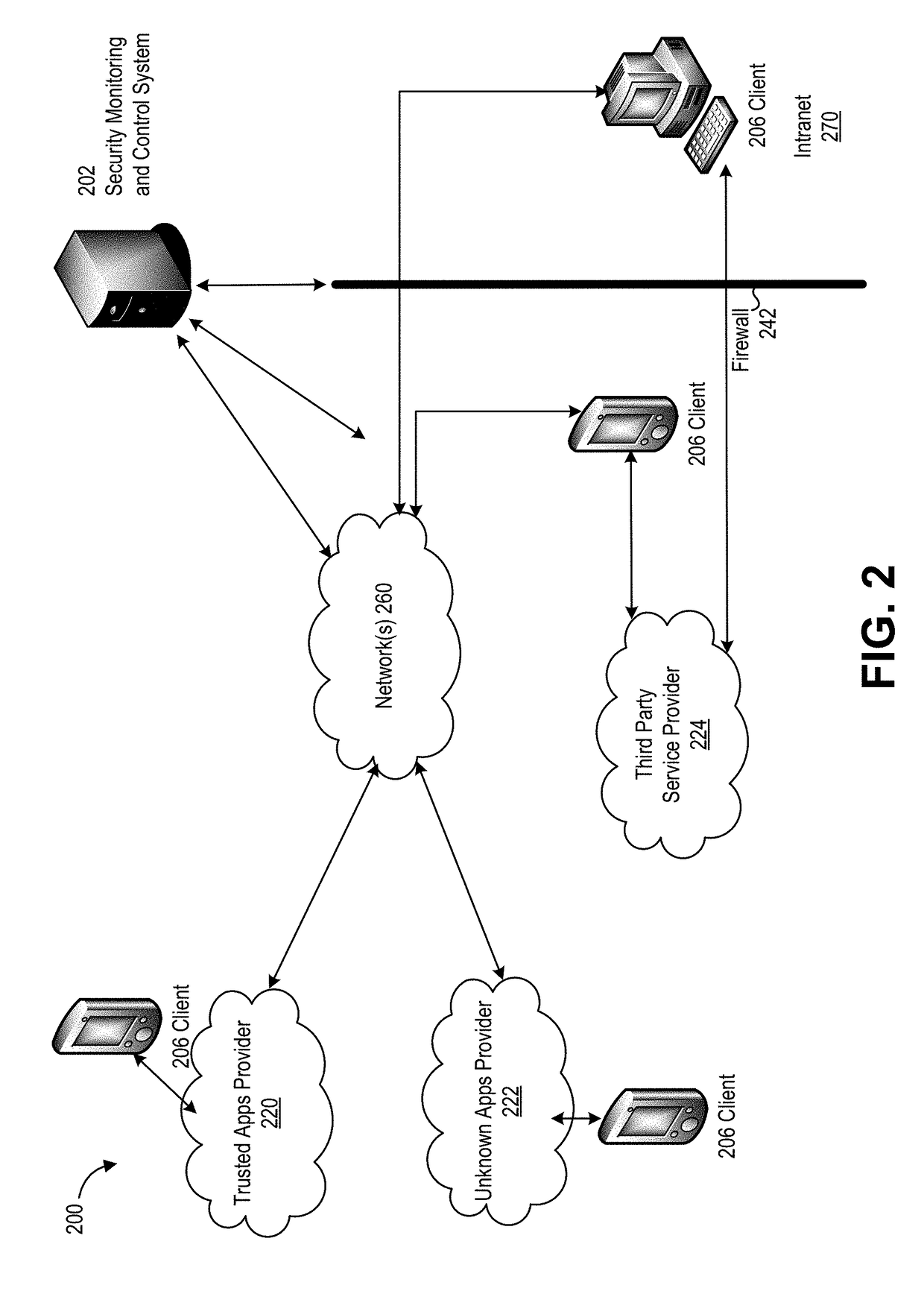
Cloud based security monitoring using unsupervised pattern recognition and deep learning
Provided are systems and methods for a cloud security system that learns patterns of user behavior and uses the patterns to detect anomalous behavior in a network. Techniques discussed herein include obtaining activity data from a service provider system. The activity data describes actions performed during use of a cloud service over a period of time. A pattern corresponding to a series of actions performed over a subset of time can be identified. The pattern can be added a model associated with the cloud service. The model represents usage of the cloud service by the one or more users. Additional activity data can be obtained from the service provider system. Using the model, a set of actions can be identified in the additional activity data that do not correspond to the model. The set of actions and an indicator that identifies the set of actions as anomalous can be output.
Owner:ORACLE INT CORP
System and method for pose-angle estimation
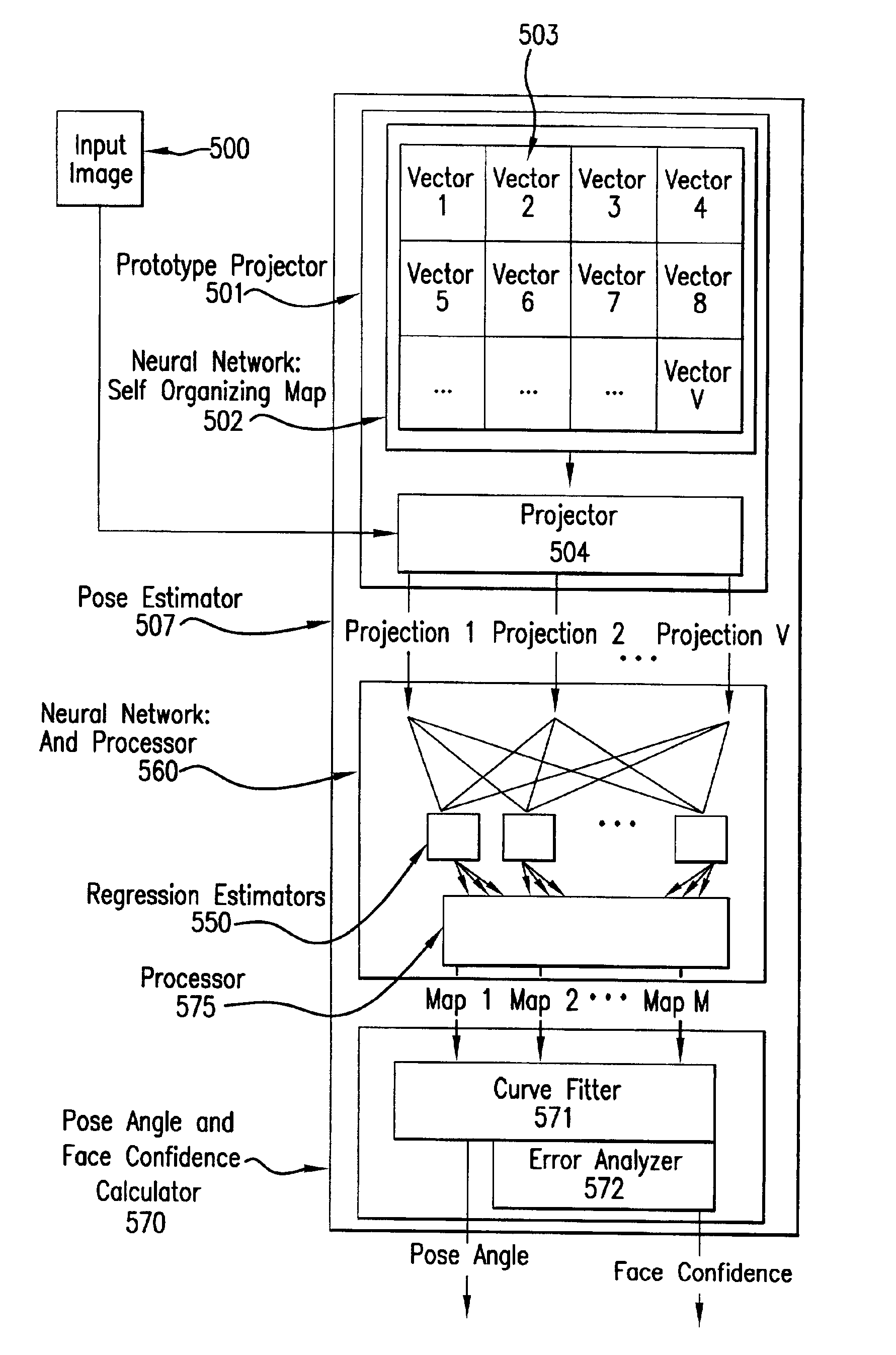
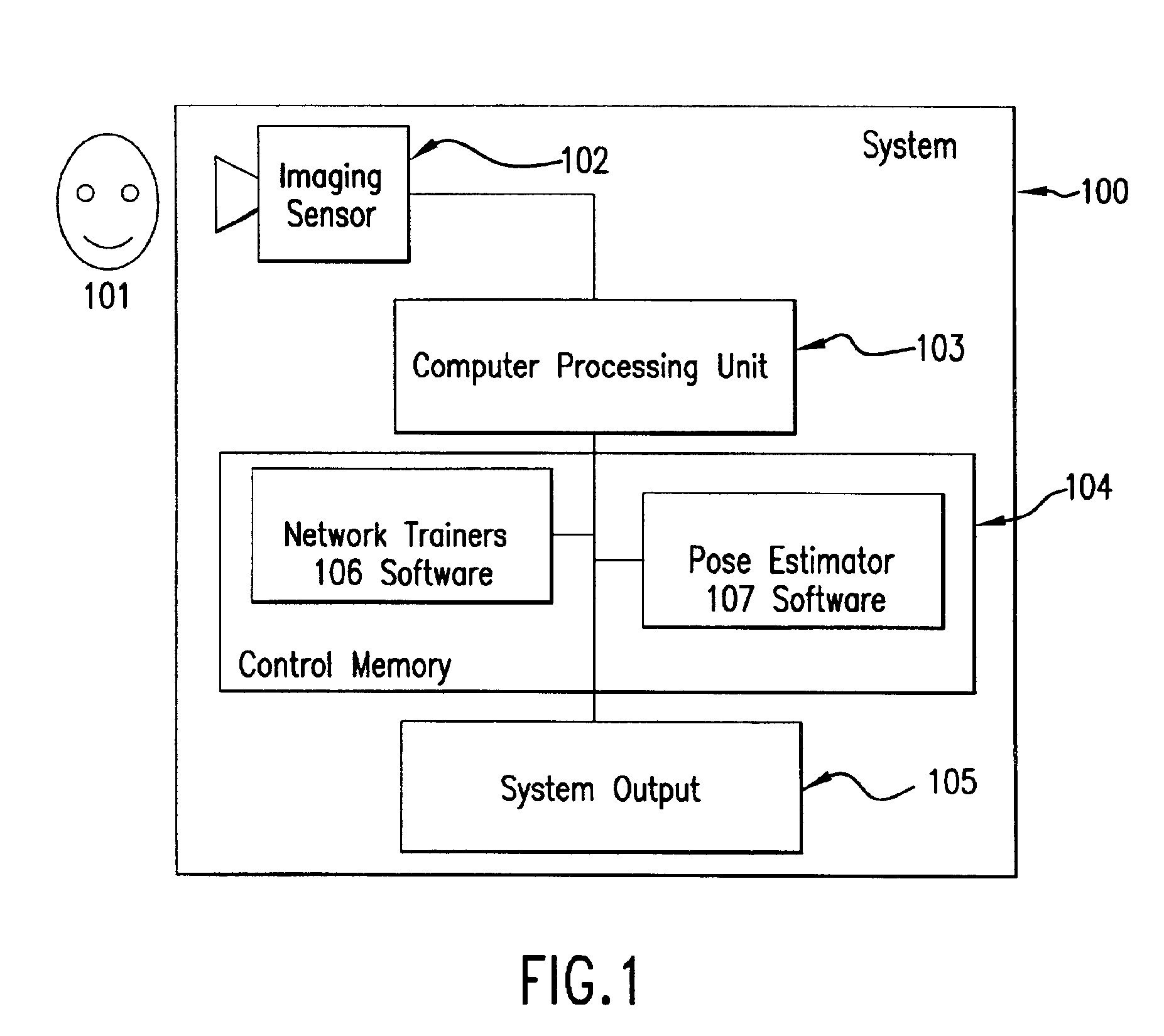
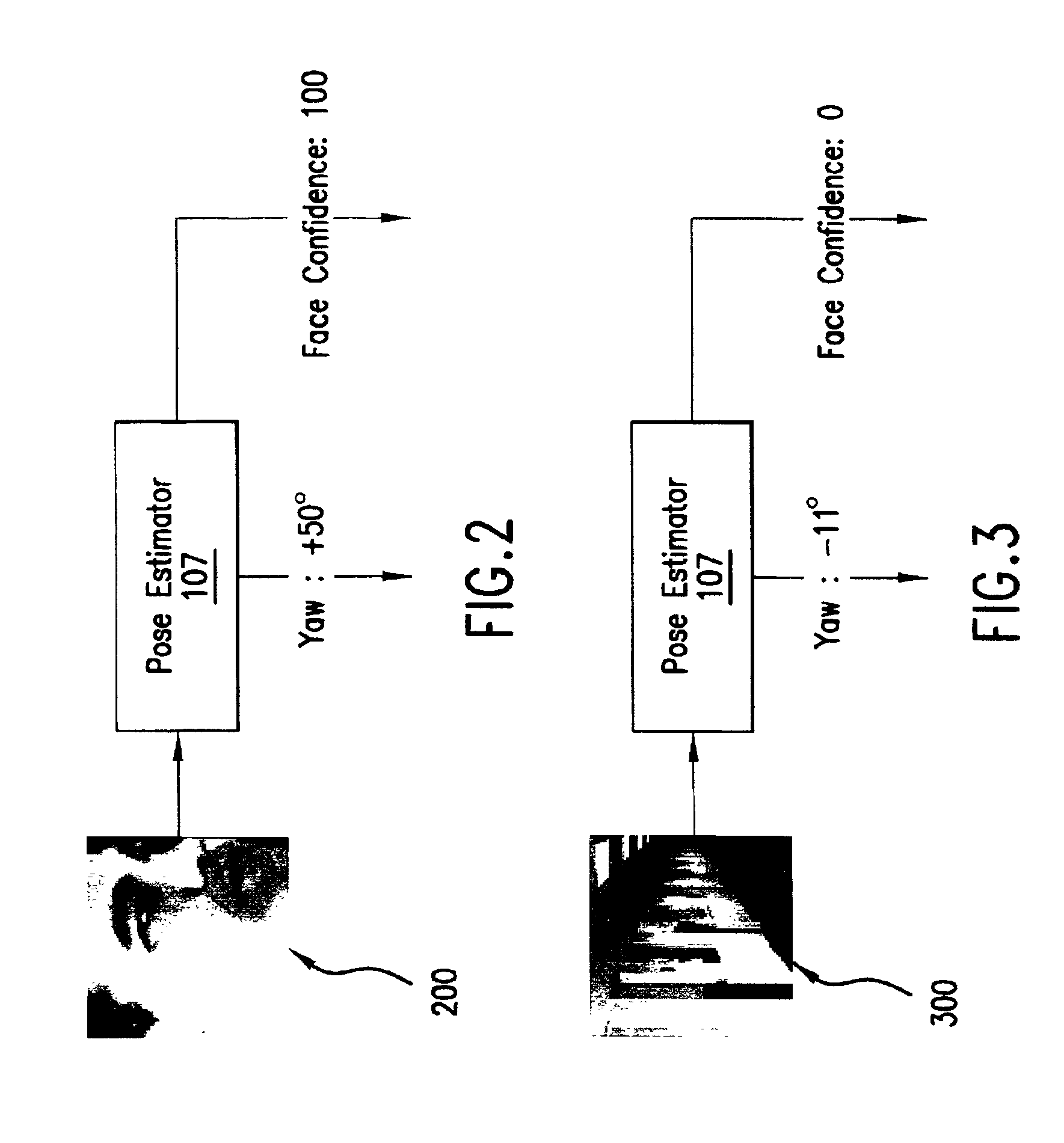
InactiveUS6959109B2Reduces input image dimensionalityElectric signal transmission systemsImage analysisPattern recognitionComputer graphics (images)
A system and method are disclosed for determining the pose angle of an object in an input image. In a preferred embodiment, the present system comprises a pose estimator having a prototype projector, a regression estimator, and an angle calculator. The prototype projector is preferably adapted to reduce the input image dimensionality for faster further processing by projecting the input pixels of the image onto a Self-Organizing Map (SOM) neural network. The regression estimator is preferably implemented as a neural network and adapted to map the projections to a pattern unique to each pose. The angle calculator preferably includes a curve fitter and an error analyzer. The curve fitter is preferably adapted to estimate the pose angle from the mapping pattern. The error analyzer is preferably adapted to produce a confidence signal representing the likelihood of the input image being a face at the calculated pose. The system also preferably includes two network trainers responsible for synthesizing the neural networks.
Owner:MORPHOTRUST USA
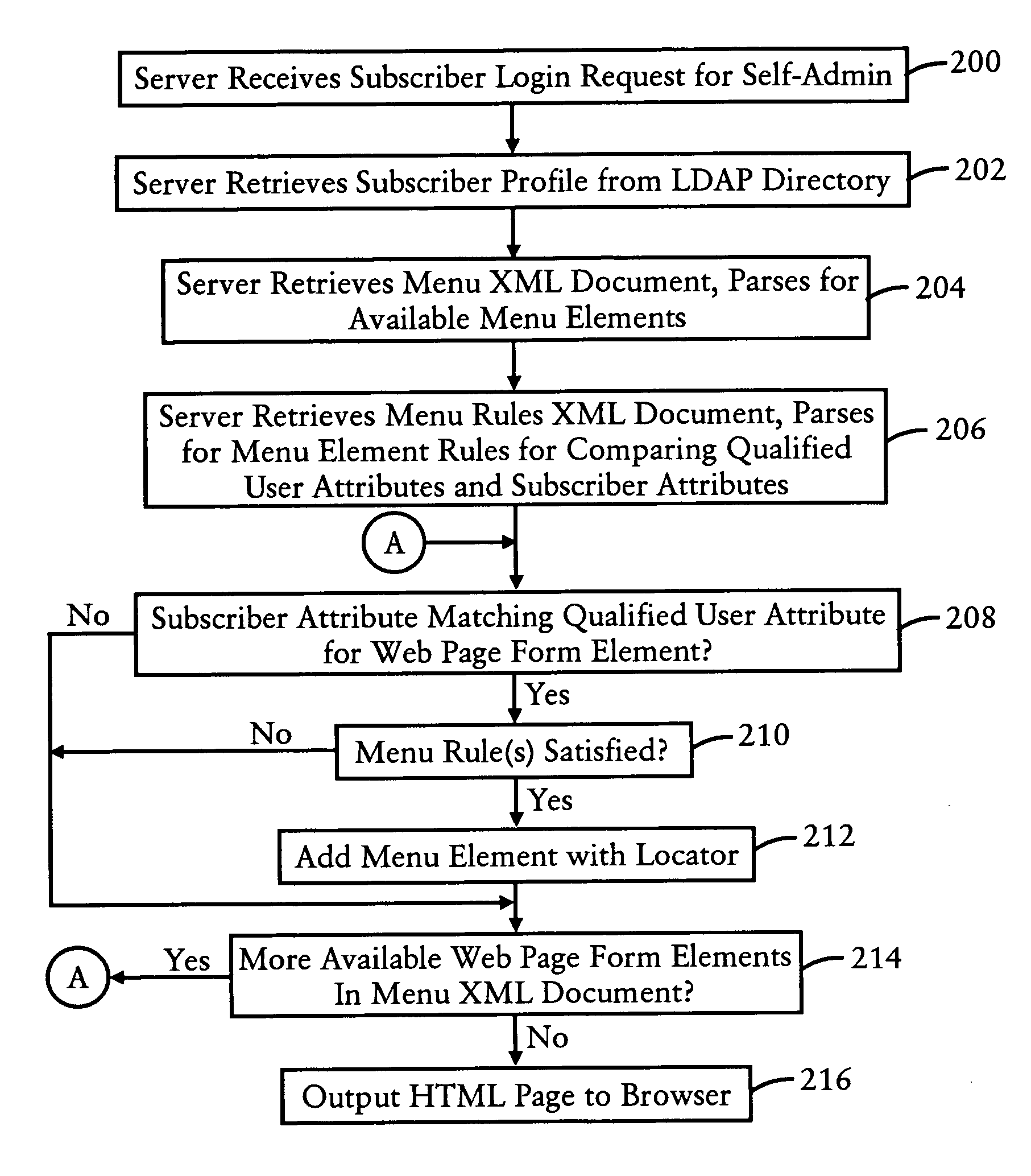
Application server configured for dynamically generating web forms based on extensible markup language documents and retrieved subscriber data
An application server, configured for dynamically generating a web page (e.g., HTML) document based on execution of XML documents, dynamically generates an HTML page having selected form elements based on a stored XML document that defines available HTML form elements and respective attributes, and based on user attributes retrieved by the application server from an open network database server (such as LDAP). The application server, in response to a request from a user, accesses an XML document configured for specifying attributes associated with the request; the XML document may specify as form elements menus that are available for generation based on qualified user attributes, or may specify HTML fields that can be generated for display on the HTML page based on the qualified user attributes. The application server also retrieves the user attributes, and dynamically generates the HTML page based on identifying the user attributes matching the qualified user attributes of the accessed XML document. Hence, HTML pages having form elements can be dynamically generated, providing personalized HTML pages without the necessity of modifying CGI scripts or stored HTML pages.
Owner:CISCO TECH INC
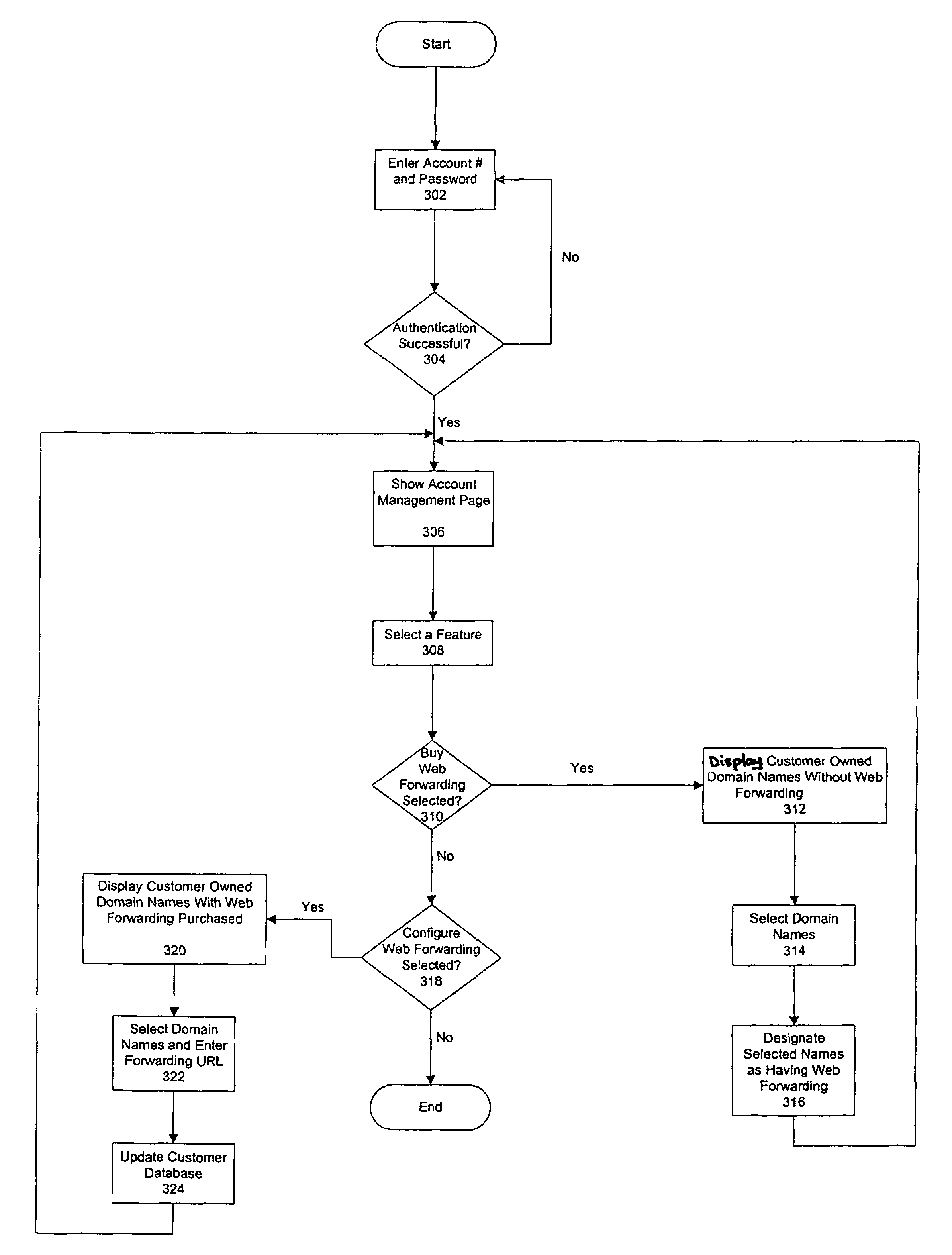
Apparatus and method for web forwarding
An apparatus and method for forwarding a web address to another web address is presented. A web forwarder receives a request destined to a first web address including at least a domain name. The web forwarder then determines a forwarding uniform resource locator (URL) that corresponds to the domain name and redirects the request to a second web address that corresponds to the forwarding URL.
Owner:NETWORK SOLUTIONS
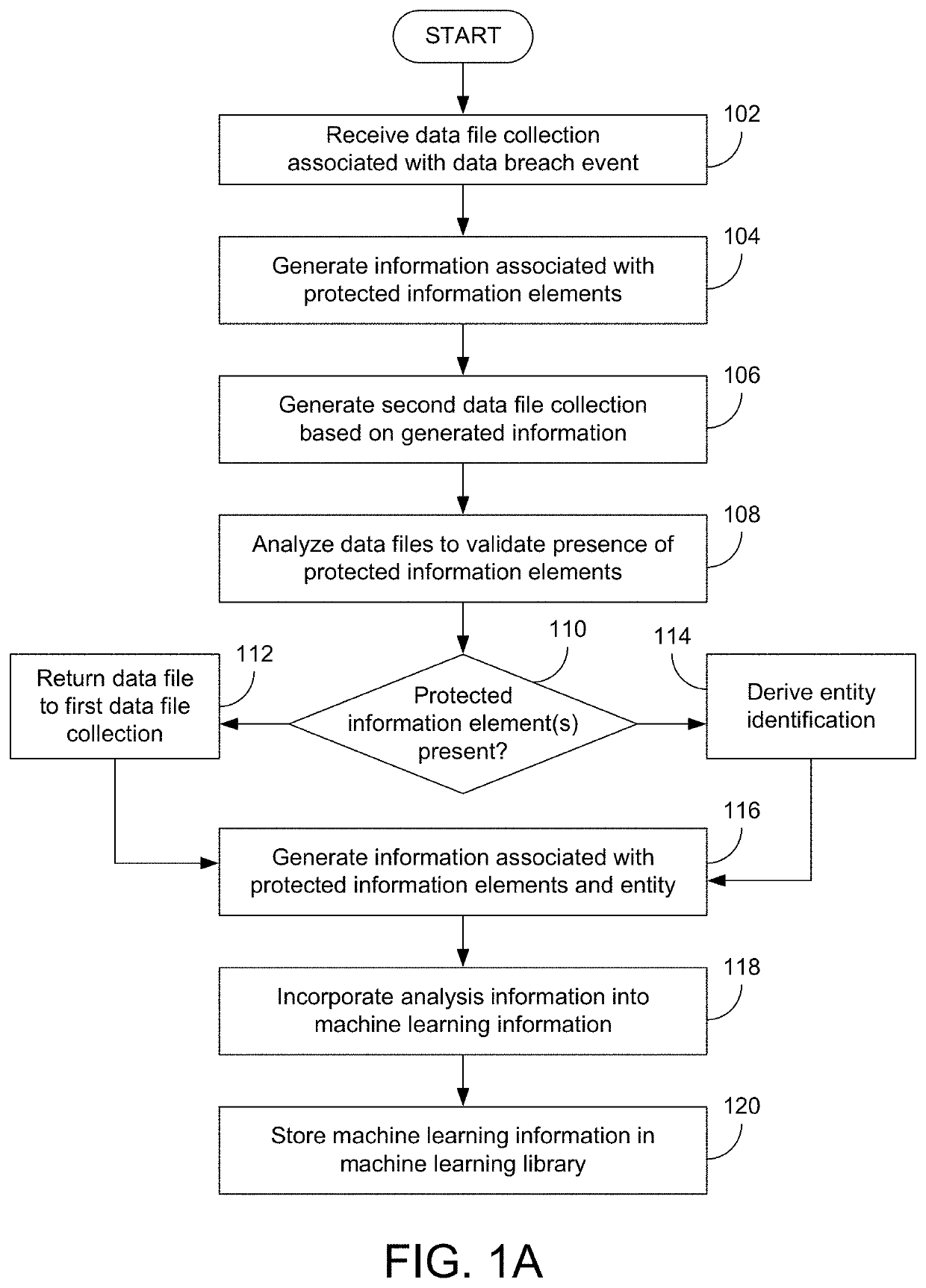
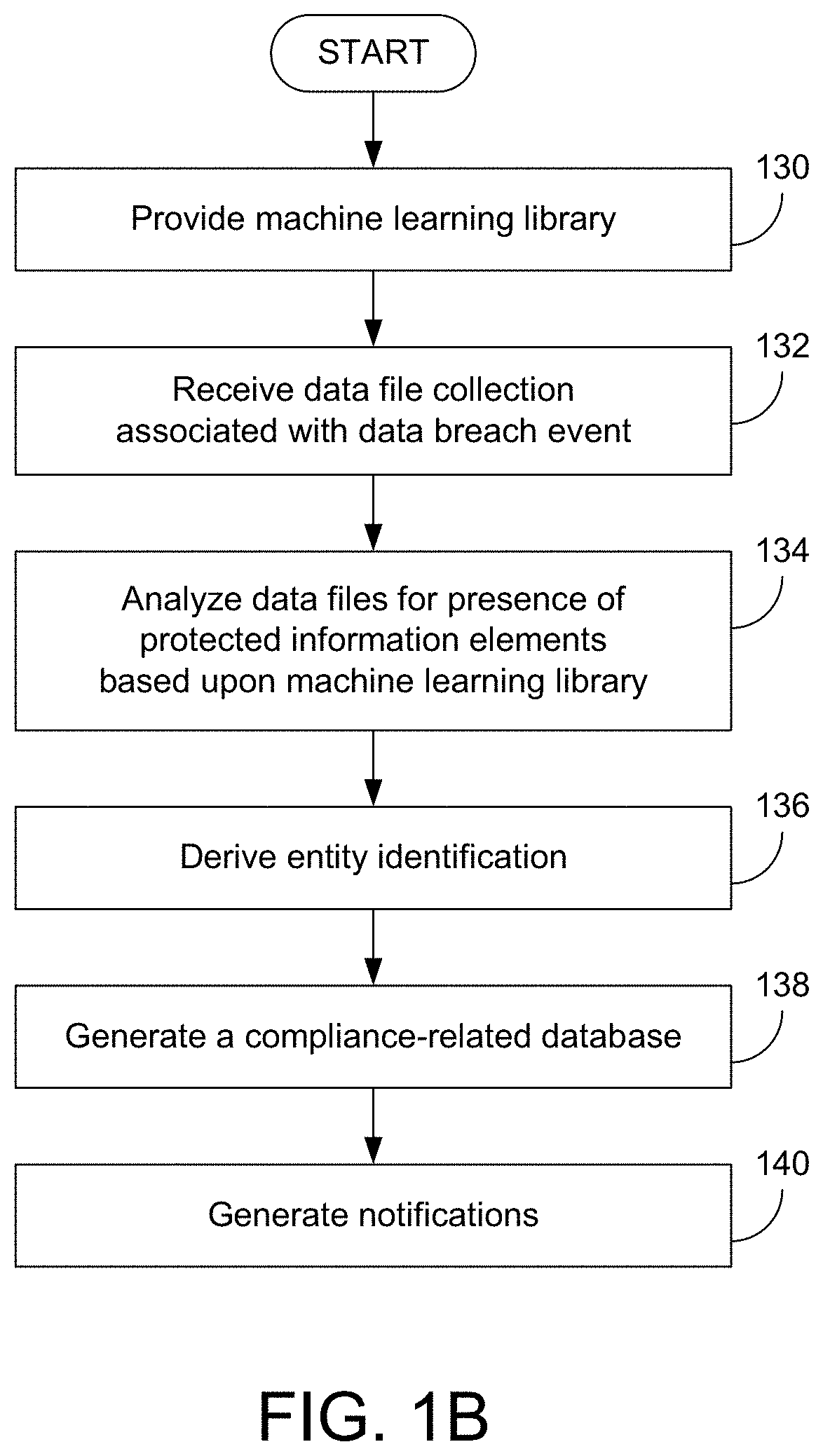
Systems And Methods For Identifying Compliance-Related Information Associated With Data Breach Events
Various examples are provided related to identification and management of compliance-related information associated with data breach events. In one example, a method includes receiving a first data file collection associated with a first data breach event; generating information associated with presence or absence of protected information elements of all or part of the first data file collection and incorporating data files including the protected information elements in a second data file collection; analyzing data files selected from the second data file collection; and incorporating the information associated with the analysis into machine learning information that may be used for subsequent analysis of data file collections.
Owner:CANOPY SOFTWARE INC
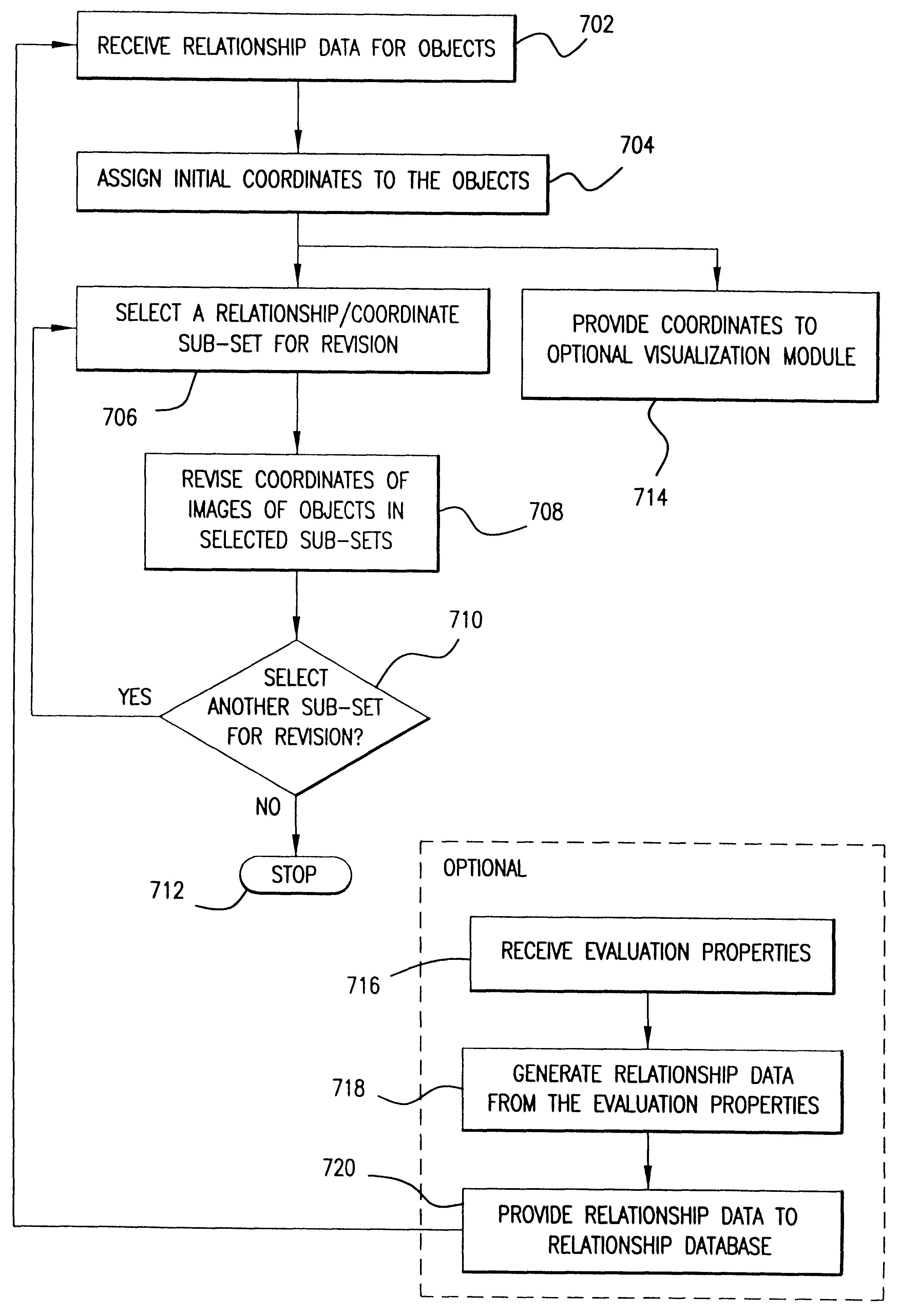
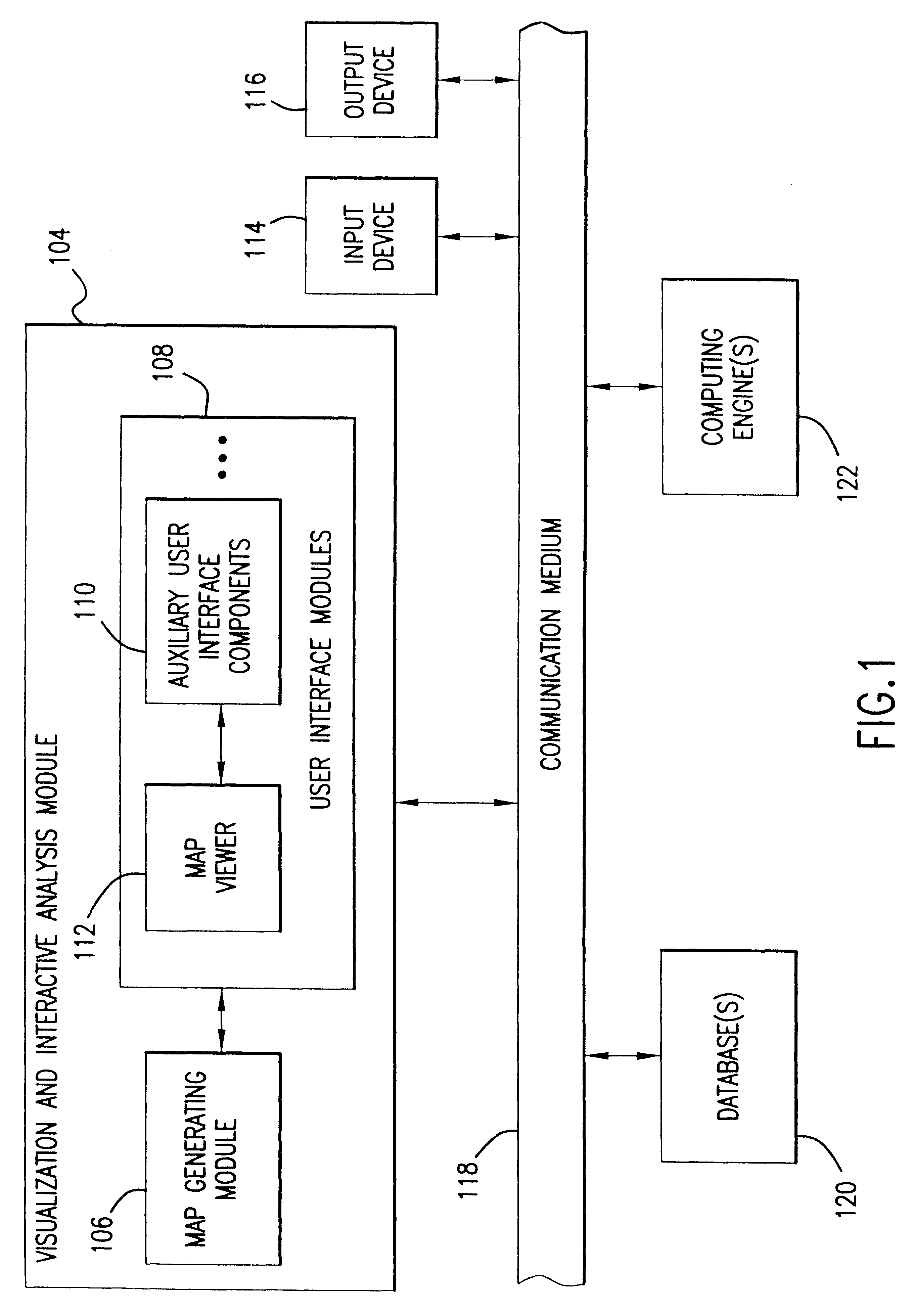
System, method, and computer program product for representing proximity data in a multi-dimensional space
A system, method and computer program product for representing precise or imprecise measurements of similarity / dissimilarity (relationships) between objects as distances between points in a multi-dimensional space that represents the objects. Self-organizing principles are used to iteratively refine an initial (random or partially ordered) configuration of points using stochastic relationship / distance errors. The data can be complete or incomplete (i.e. some relationships between objects may not be known), exact or inexact (i.e. some or all of the relationships may be given in terms of allowed ranges or limits), symmetric or asymmetric (i.e. the relationship of object A to object B may not be the same as the relationship of B to A) and may contain systematic or stochastic errors. The relationships between objects may be derived directly from observation, measurement, a priori knowledge, or intuition, or may be determined indirectly using any suitable technique for deriving proximity (relationship) data.
Owner:3 DIMENSIONAL PHARMA
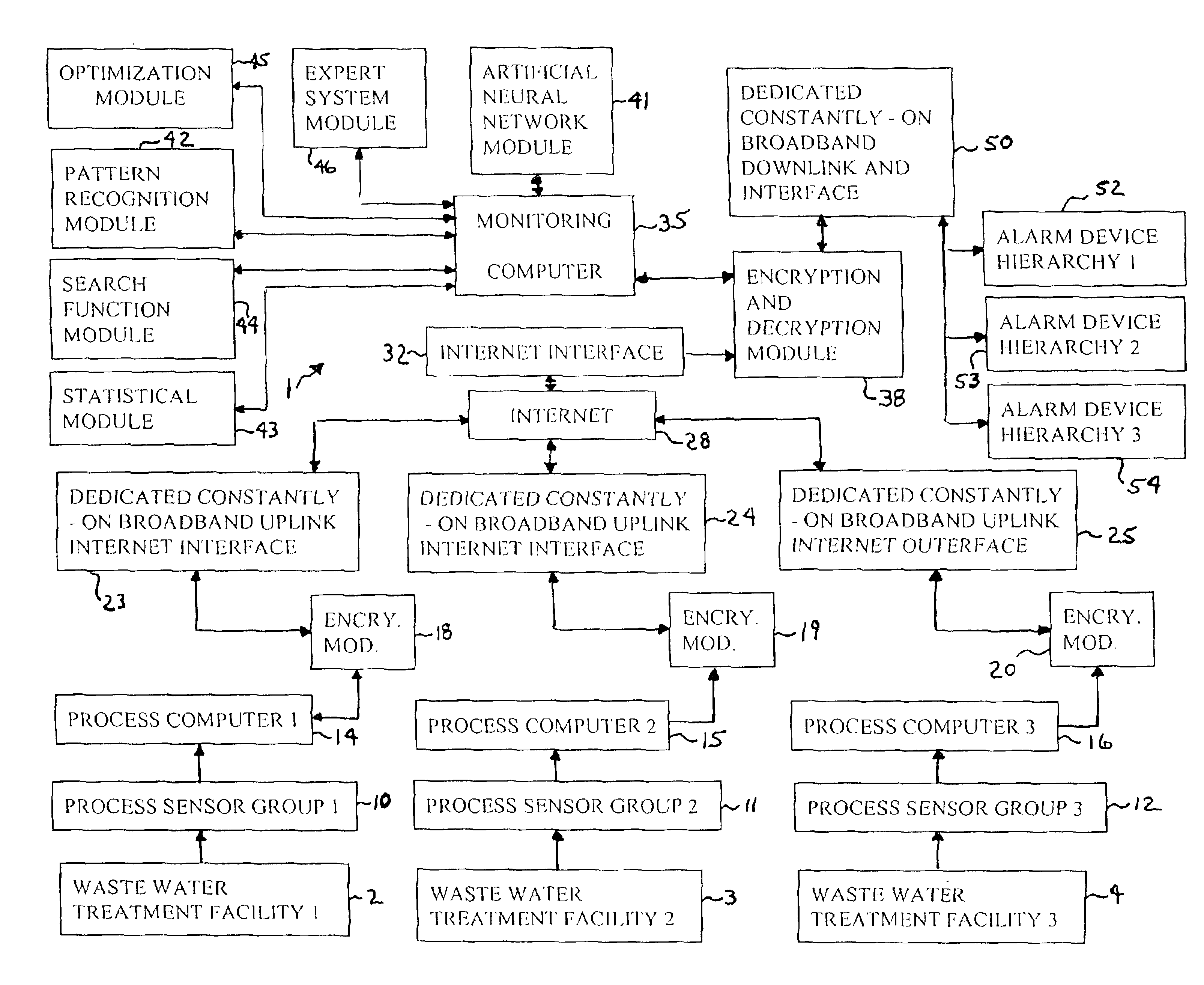
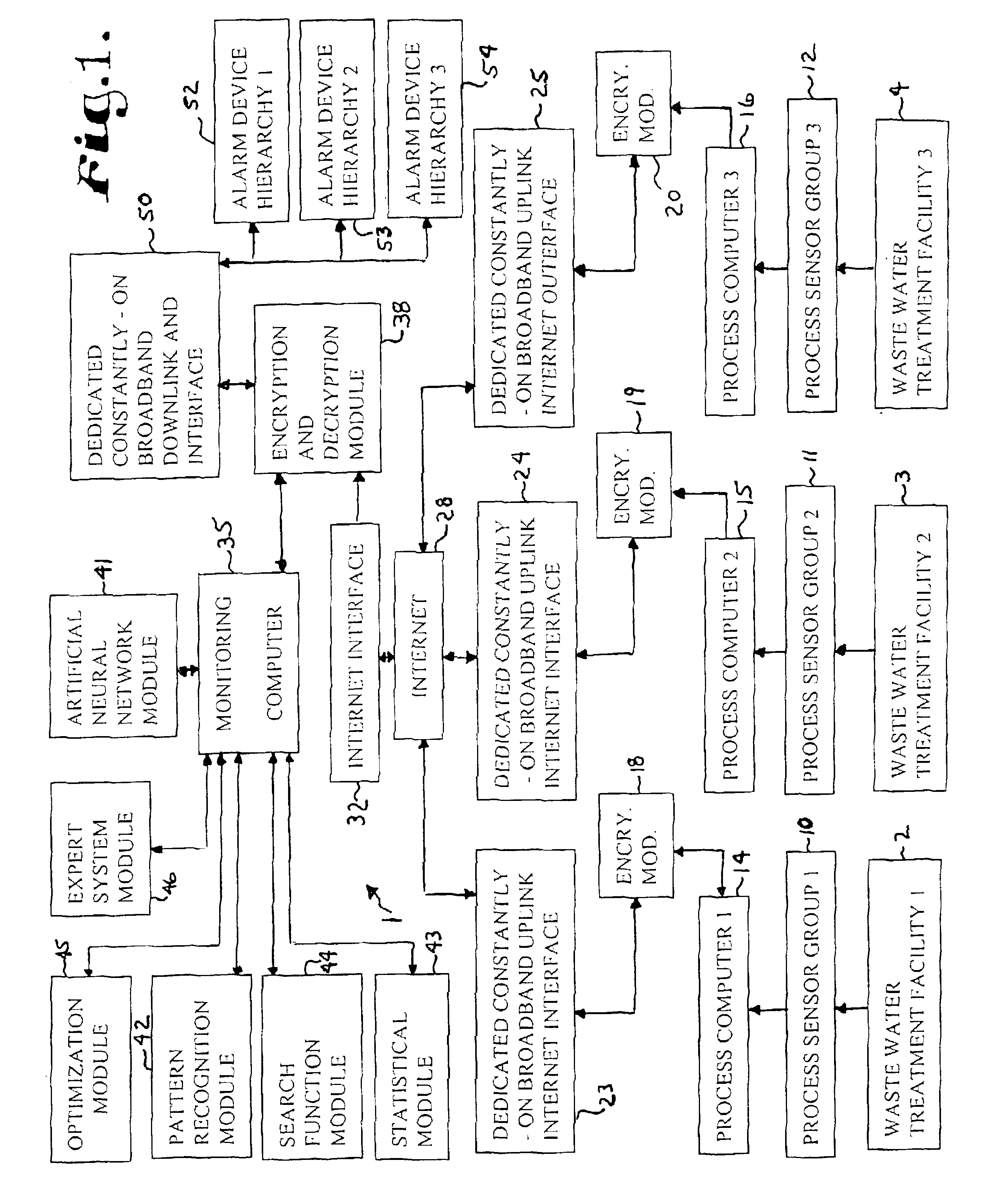
Water treatment monitoring system
InactiveUS6845336B2Reduce severityBackingTesting/calibration apparatusElectrical testingReal-time dataAnalysis data
A computer system linked by the internet to various remote waste water treatment facilities. The system receives real-time data from the facilities and analyzing the data to determine likely operational upsets and future effluent water quality. The computer system sends signals to a hierarchy of parties depending on the severity of predicted upsets problems and events. The computer also provides a probability distribution of such upsets and water quality and recommendations as how to adjust facility operating parameters to avoid or reduce the upsets to acceptable parameters and maintain effluent water quality parameters within preselected limits.
Owner:NEOCHLORIS
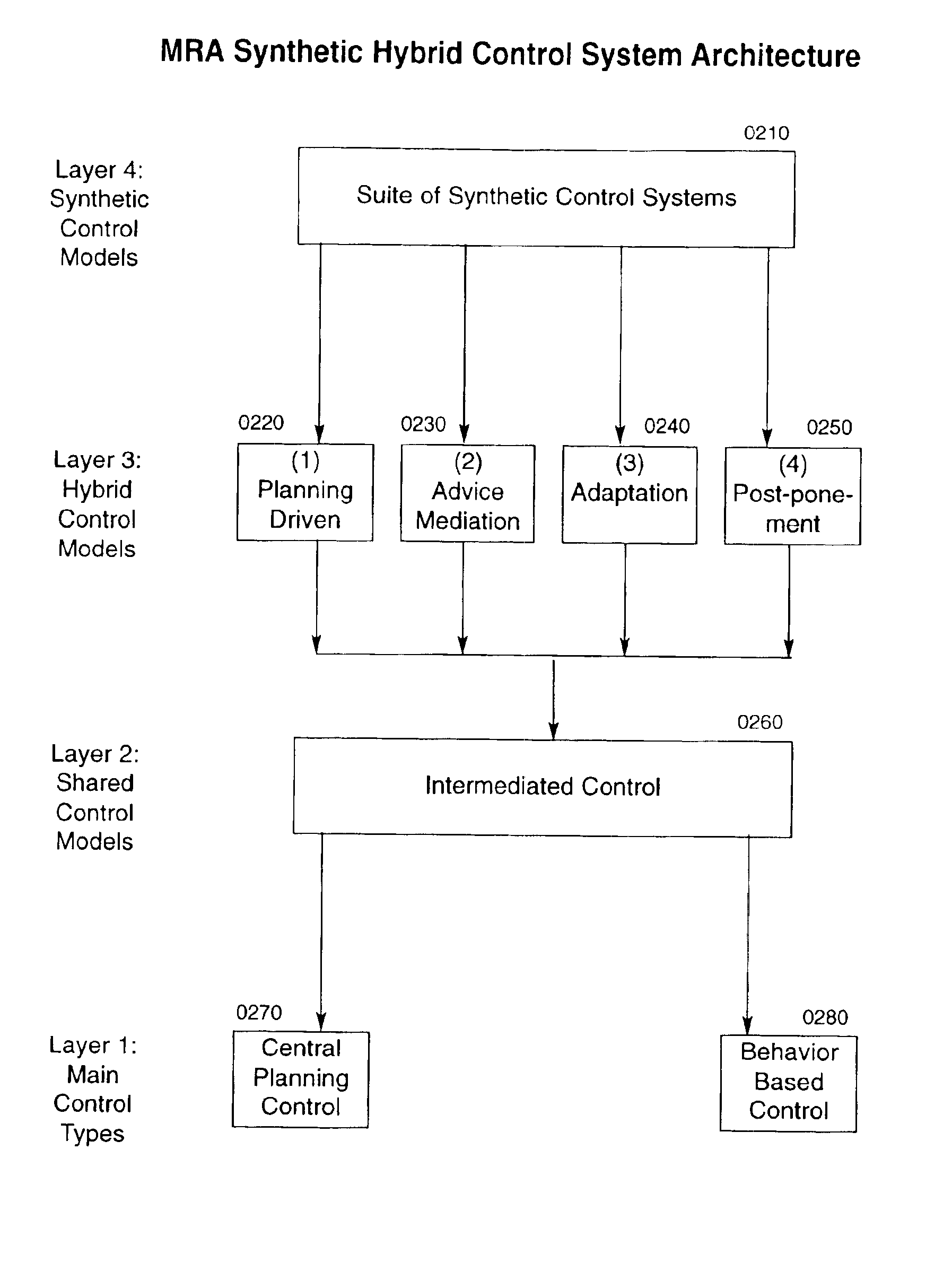
System, method and apparatus for organizing groups of self-configurable mobile robotic agents in a multi-robotic system
InactiveUS6904335B2Severe resource restrictionComplex behaviorProgramme-controlled manipulatorAutonomous decision making processRobotic systemsMultirobot systems
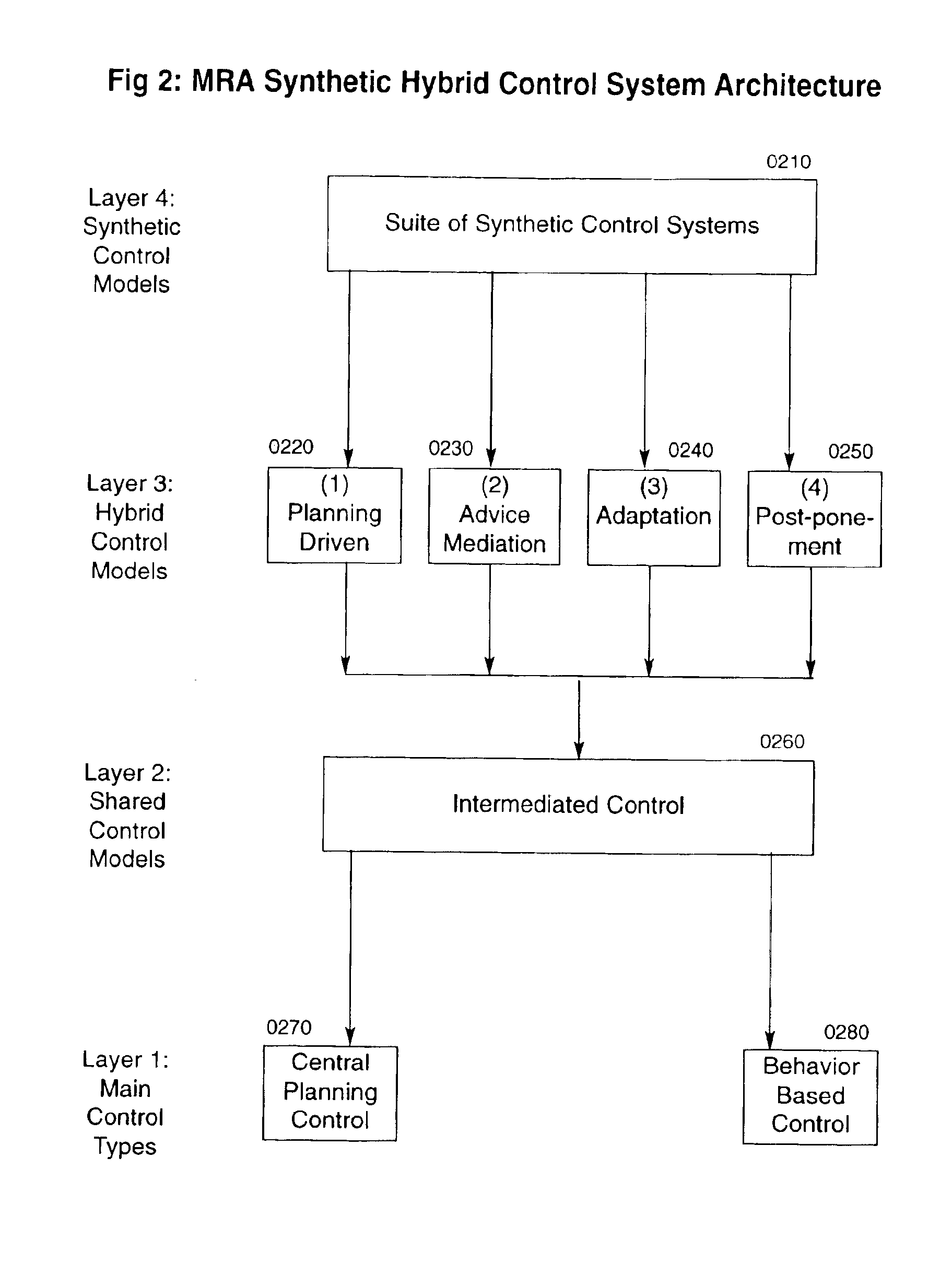
A system of self-organizing mobile robotic agents (MRAs) in a multi-robotic system (MRS) is disclosed. MRAs cooperate, learn and interact with the environment. The system uses various AI technologies including genetic algorithms, genetic programming and evolving artificial neural networks to develop emergent dynamic behaviors. The collective behaviors of autonomous intelligent robotic agents are applied to numerous applications. The system uses hybrid control architectures. The system also develops dynamic coalitions of groups of autonomous MRAs for formation and reformation in order to perform complex tasks.
Owner:SOLOMON RES
Signal processing method and system for noise removal and signal extraction
InactiveUS7519488B2Noise figure or signal-to-noise ratio measurementCharacter and pattern recognitionTime domainDecomposition
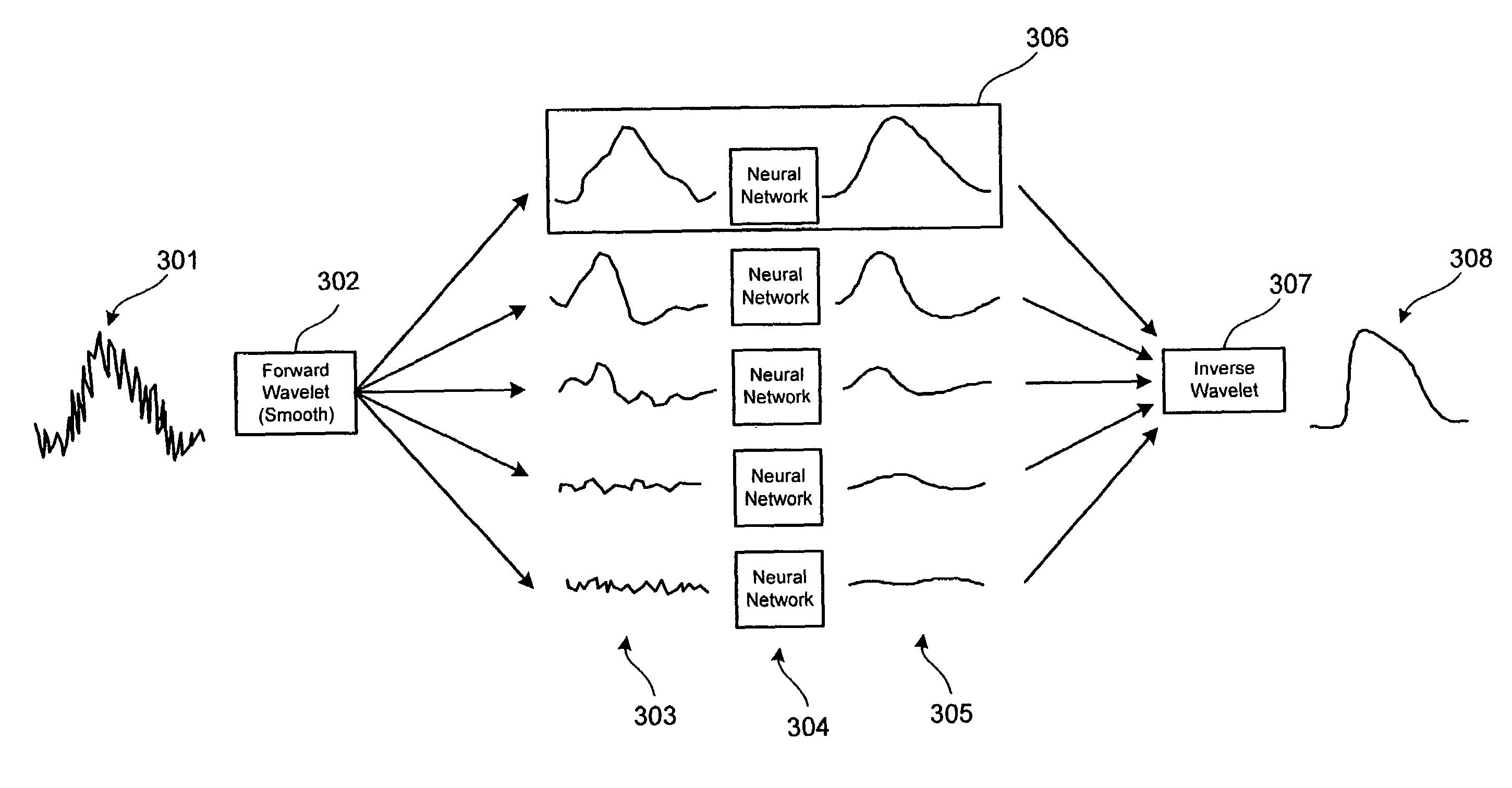
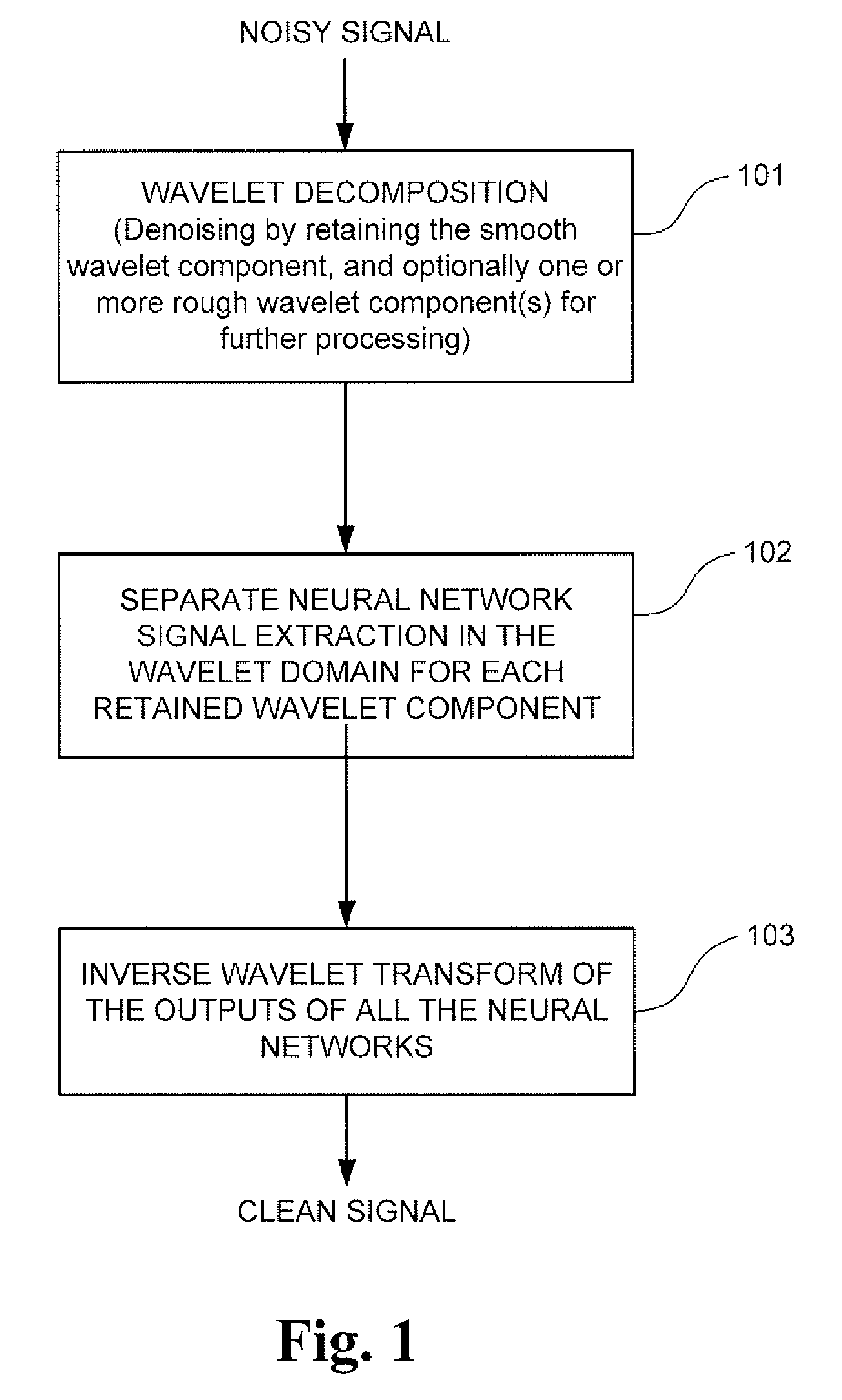
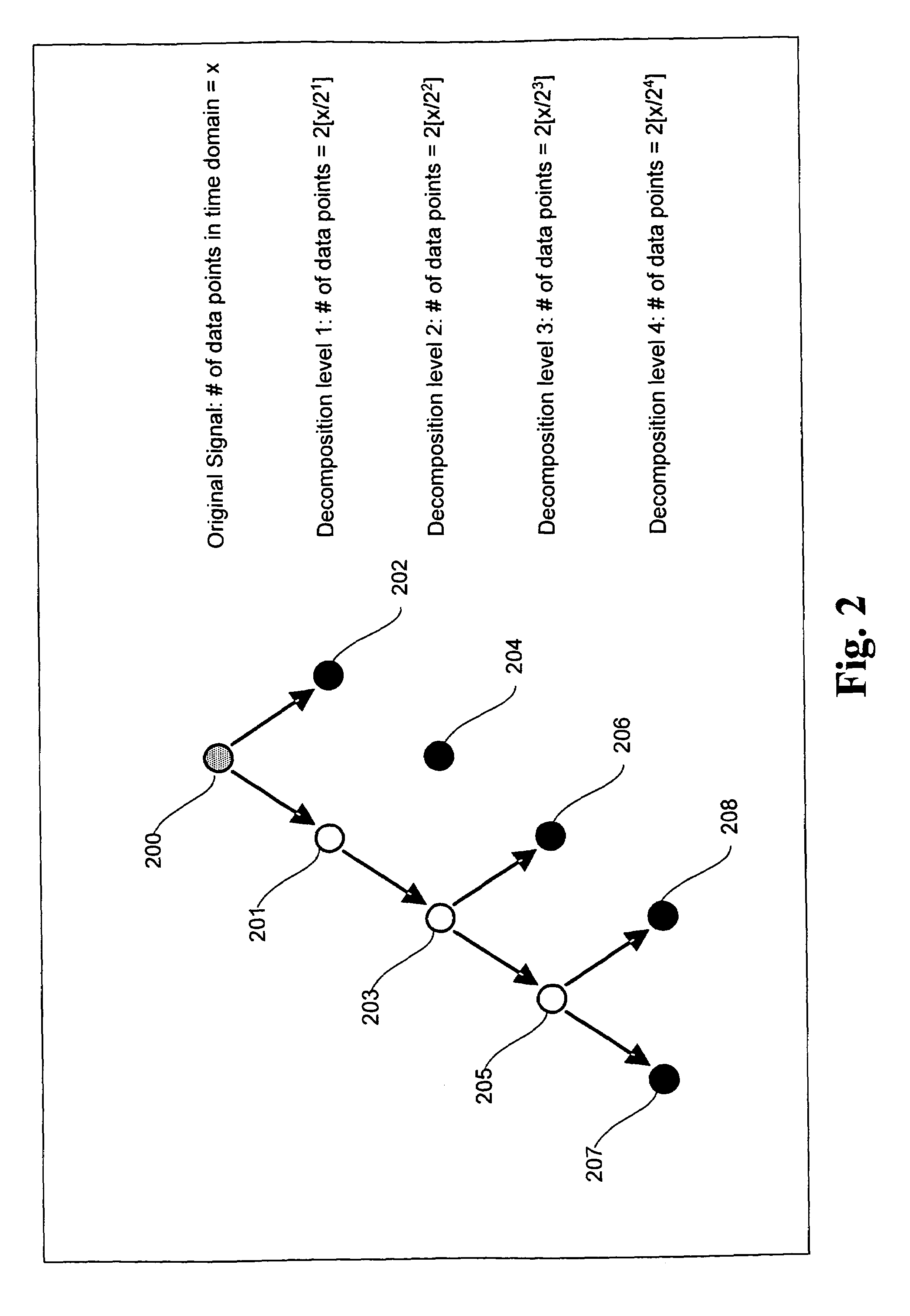
A signal processing method and system combining smooth level wavelet pre-processing together with artificial neural networks all in the wavelet domain for signal denoising and extraction. Upon receiving a signal corrupted with noise, an n-level decomposition of the signal is performed using a discrete wavelet transform to produce a smooth component and a rough component for each decomposition level. The nth level smooth component is then inputted into a corresponding neural network pre-trained to filter out noise in that component by pattern recognition in the wavelet domain. Additional rough components, beginning at the highest level, may also be retained and inputted into corresponding neural networks pre-trained to filter out noise in those components also by pattern recognition in the wavelet domain. In any case, an inverse discrete wavelet transform is performed on the combined output from all the neural networks to recover a clean signal back in the time domain.
Owner:LAWRENCE LIVERMORE NAT SECURITY LLC
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com