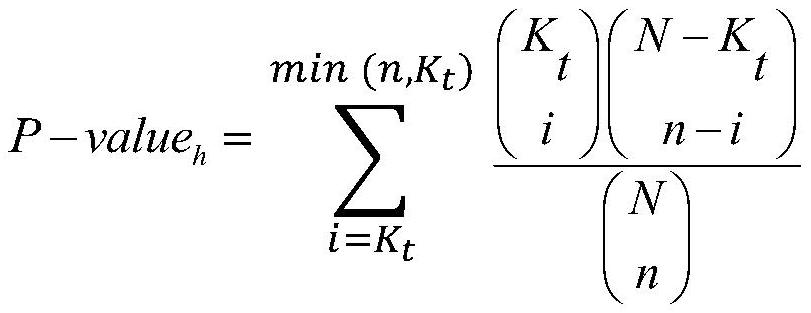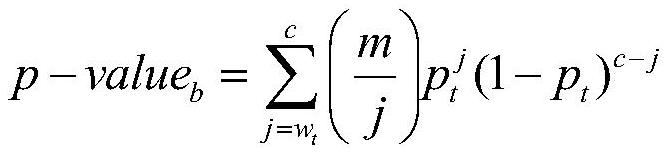Patents
Literature
33 results about "Gene association" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Genetic association is a test of a relationship between a particular phenotype and a specific allele of a gene.
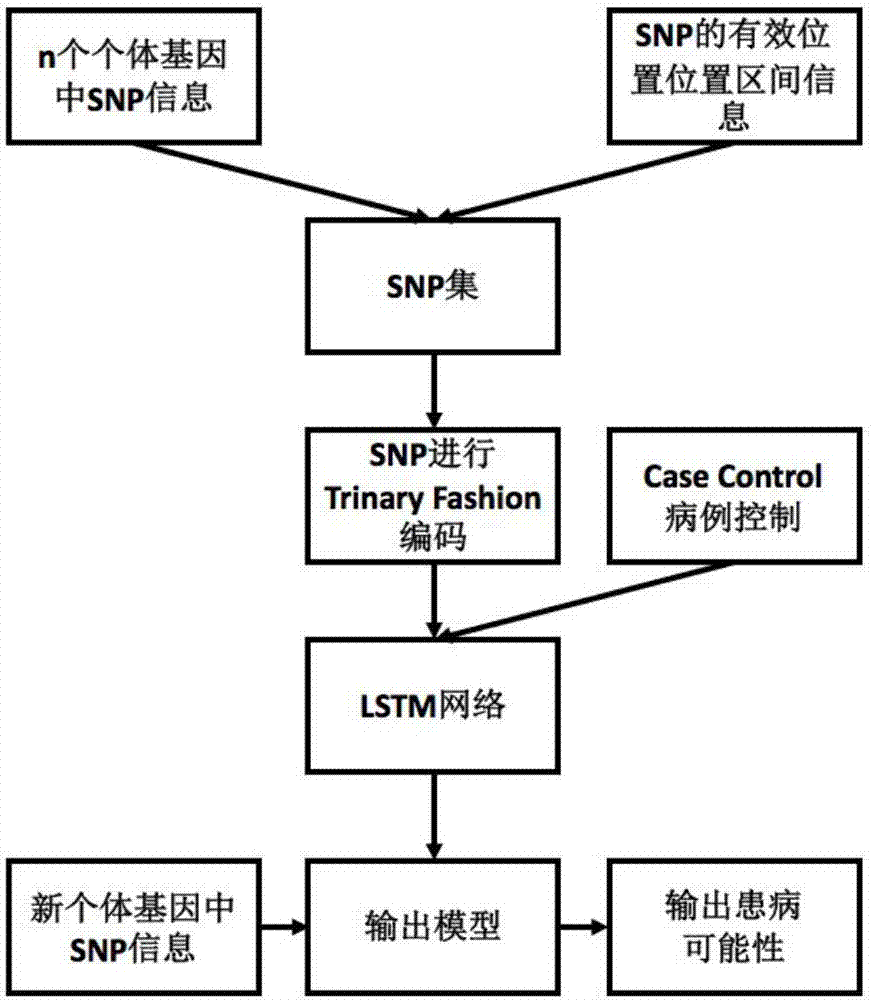
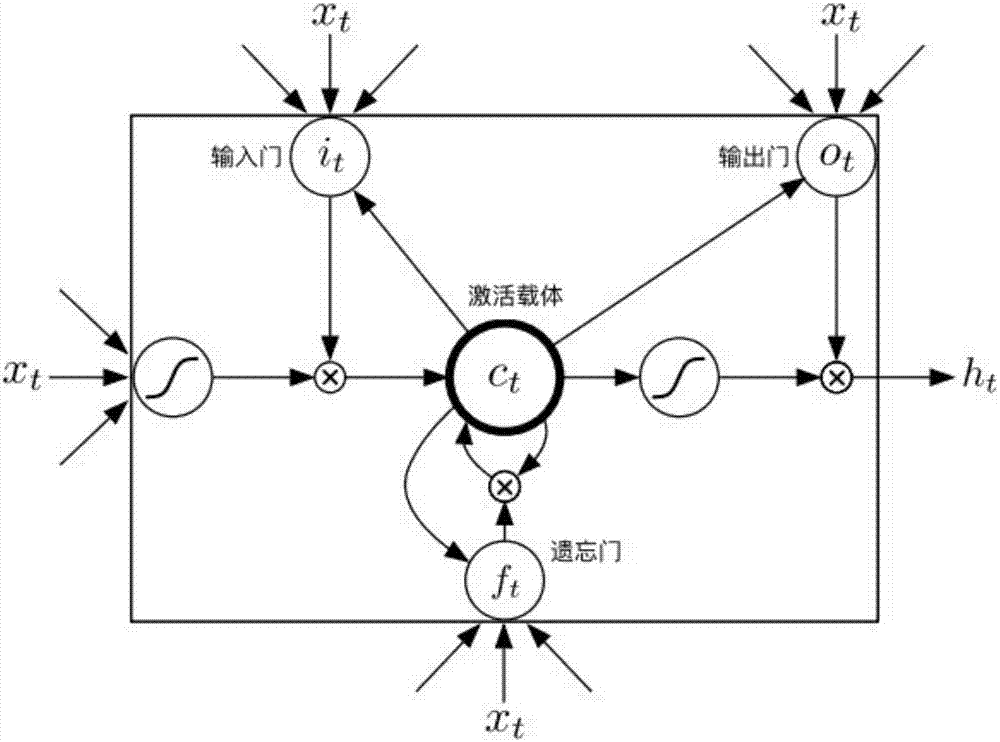
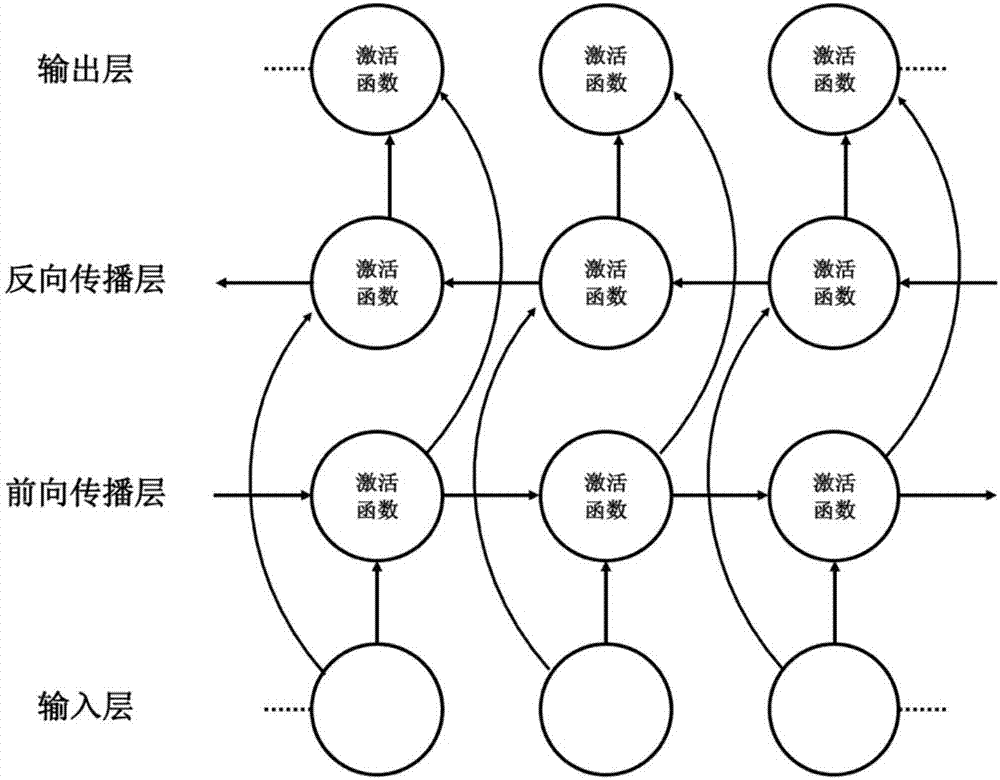
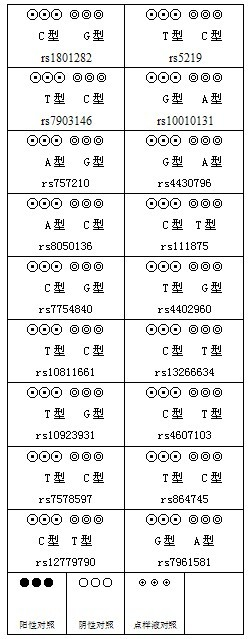
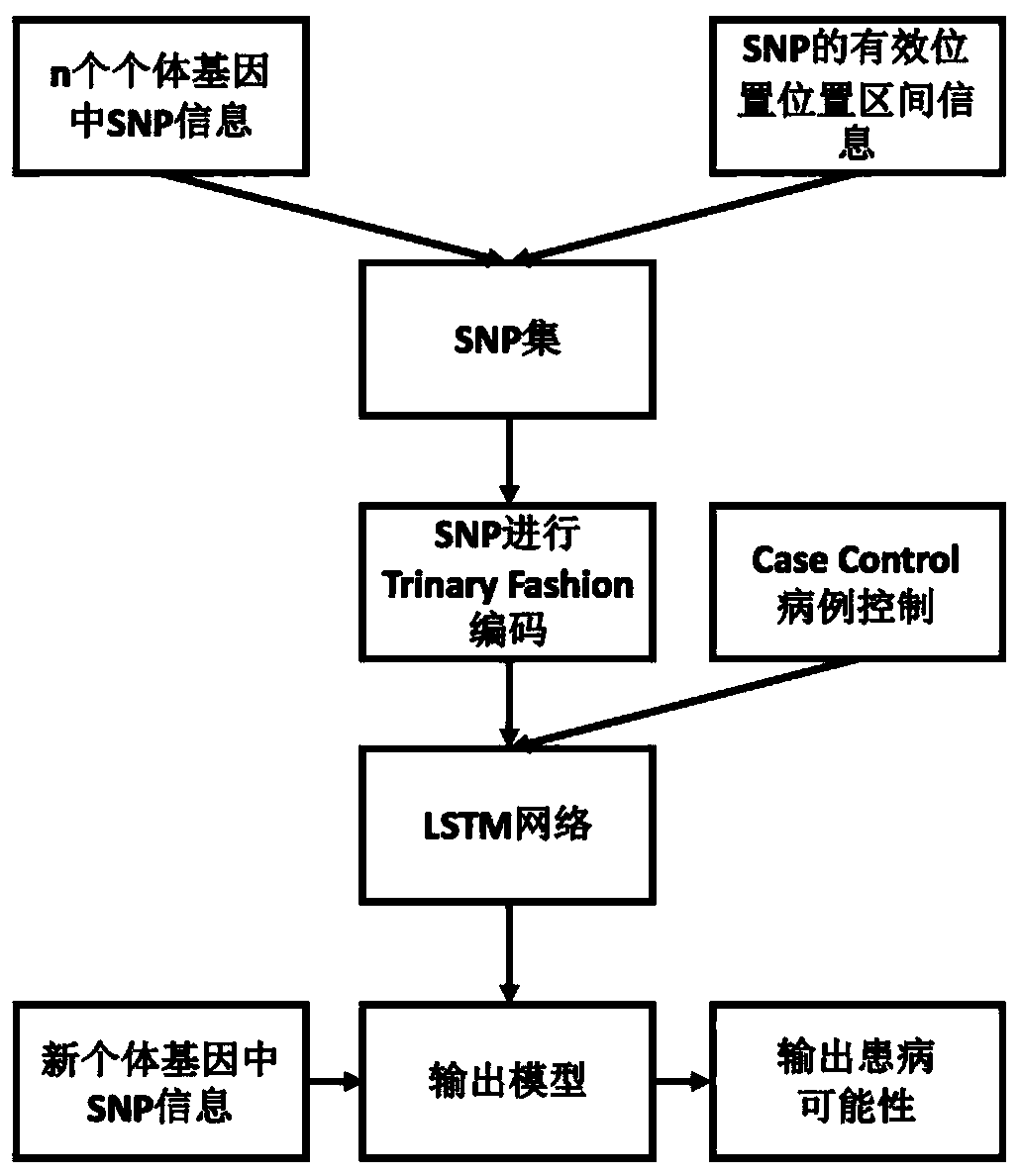
Method for gene association analysis on basis of deep learning algorithm
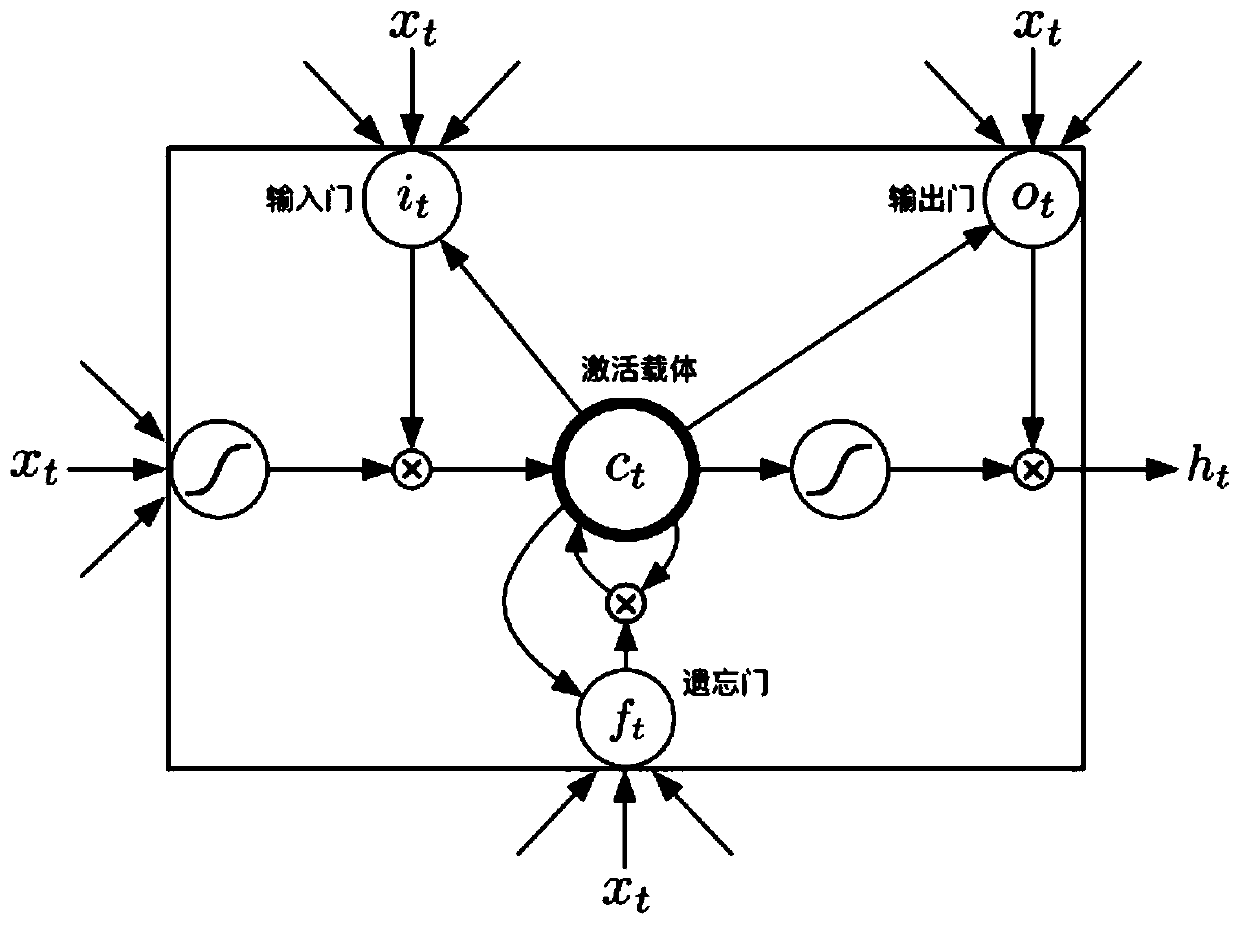
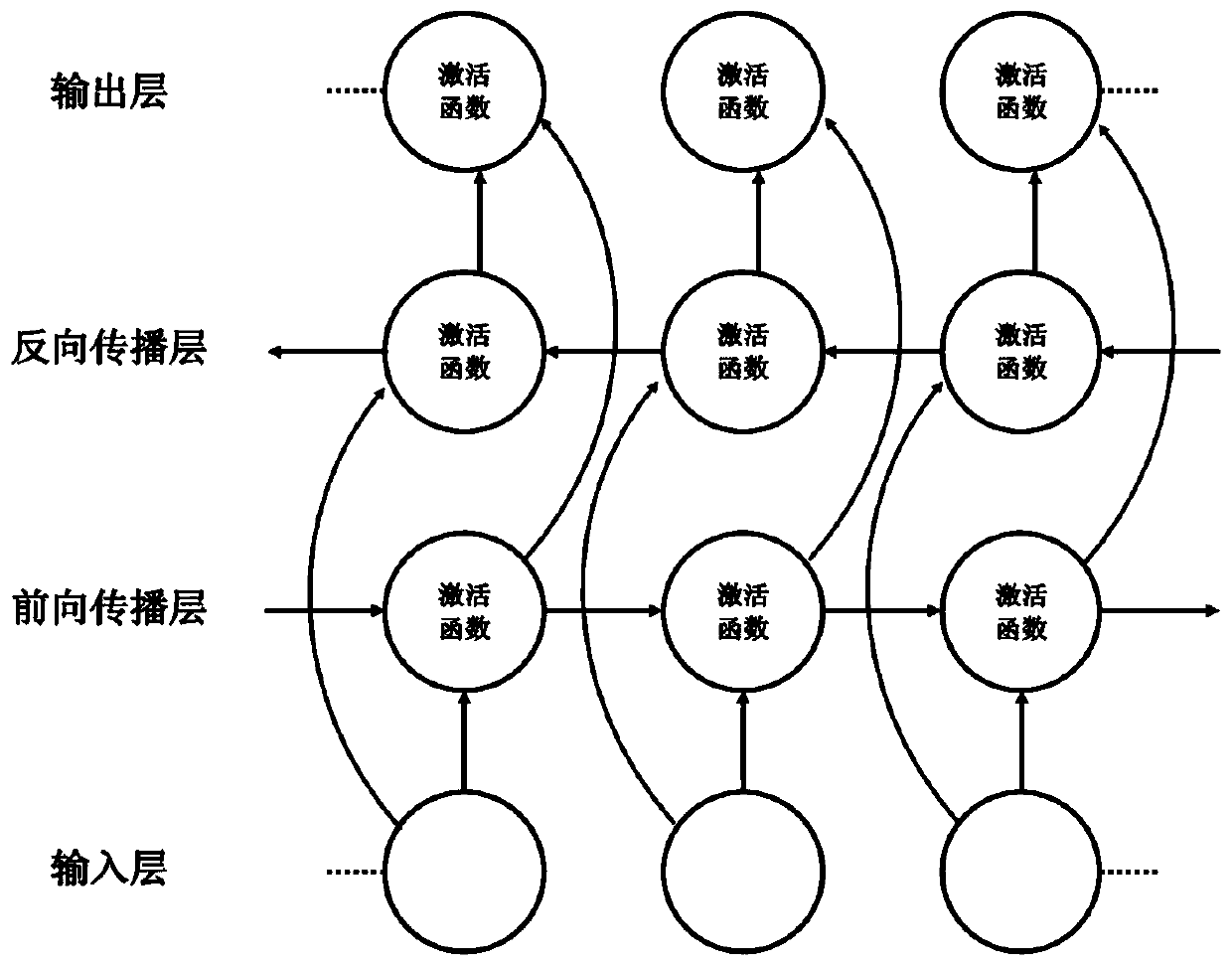
The invention discloses a method for gene association analysis on basis of a deep learning algorithm. The method based on SNP (single nucleotide polymorphism) set analysis needs to take SNP information, which comes from different positions of the same individual but is related, as a reference, and SNP of the individual is divided into multiple units according to existing biological knowledge. Firstly, all SNP is divided into multiple SNP sets on the whole chromosome level according to related knowledge of biology such as the principles approaching genomic characteristics. After division ends, each SNP set is input into an established bidirectional LSTM (long short-term memory) network, the network is a circulating neural network, and the state of the network contains old information of the last moment and also is a basis of weight change of the next moment. After LSTM network learning is completed, the attention degree required for input data can be output through network calculation. The method has better sensitivity and specificity and a new field is explored for development and research of the clinical medicine, genetic epidemiology and preventive medicine.
Owner:HANGZHOU DIANZI UNIV
Gene chip for detecting mutation of 18 loci of susceptibility genes of type 2 diabetes
ActiveCN101956017AGood consistency of fluorescence intensityEasy to operateMicrobiological testing/measurementFluorescence/phosphorescencePositive controlQuality control system
The invention discloses a gene chip for detecting mutation of 18 loci of susceptibility genes of type 2 diabetes. The chip takes probes shown as SEQ ID No.1-36 which are fixed on a solid phase carrier as a detection system, and further comprises positive control, negative control and parallel control which form a quality control system package. The invention also discloses supporting primers for the detection of the gene chip. The primers can perform polymerase chain reaction (PCR) at the same annealing temperature, amplify and accomplish Cy3 marking at the same time, and perform hybridization reaction at one time to detect the 18 gene loci. The gene chip has the advantages of accurate detection result, fastness and high efficiency, can systematically screen the mutation of the 18 loci ofthe susceptibility genes of the type 2 diabetes which are reported at home and abroad at present, and is suitable for large-sample low-cost association studies on the susceptibility genes of the type2 diabetes.
Owner:GUANGZHOU IMPROVE MEDICAL TECH CO LTD +1
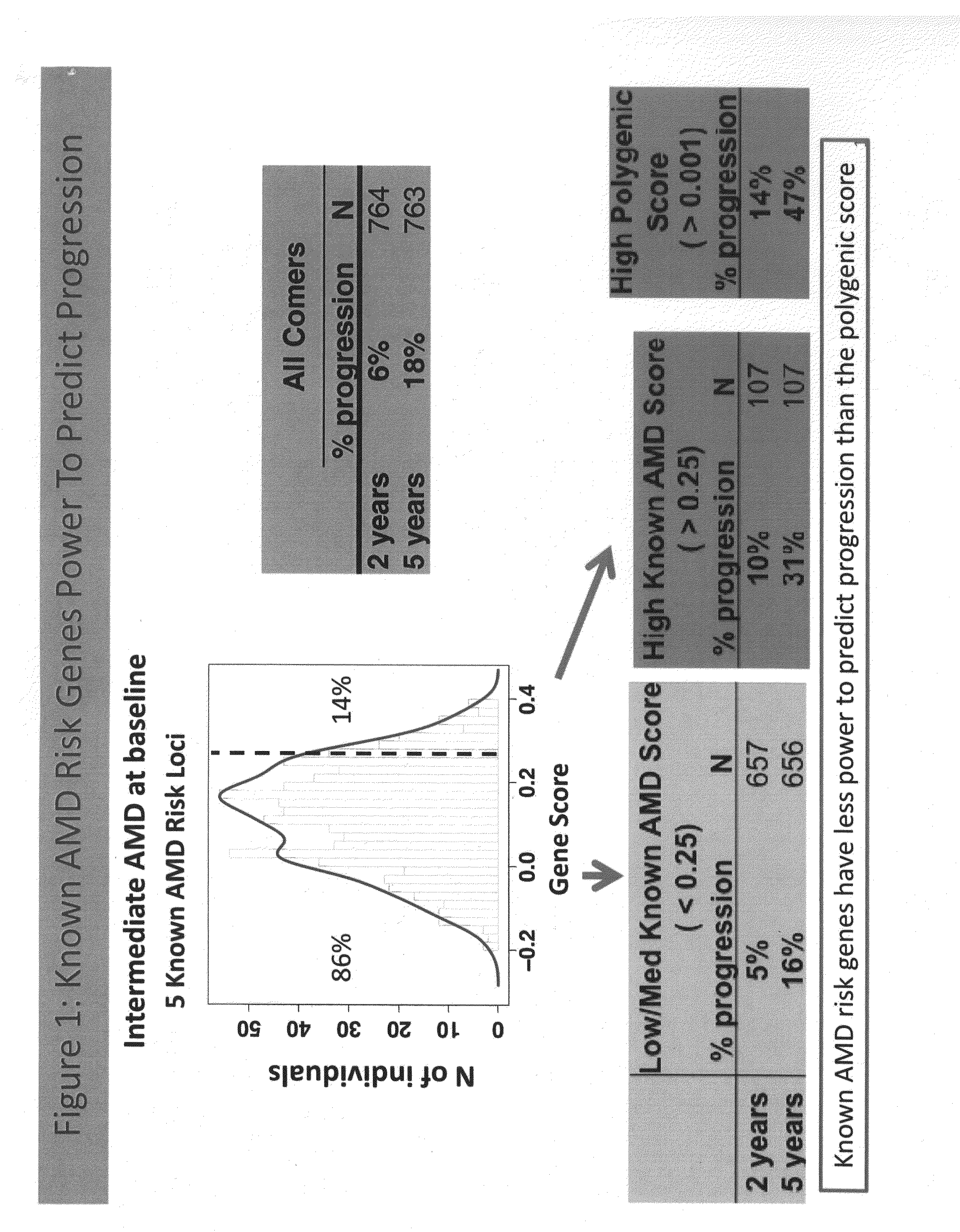
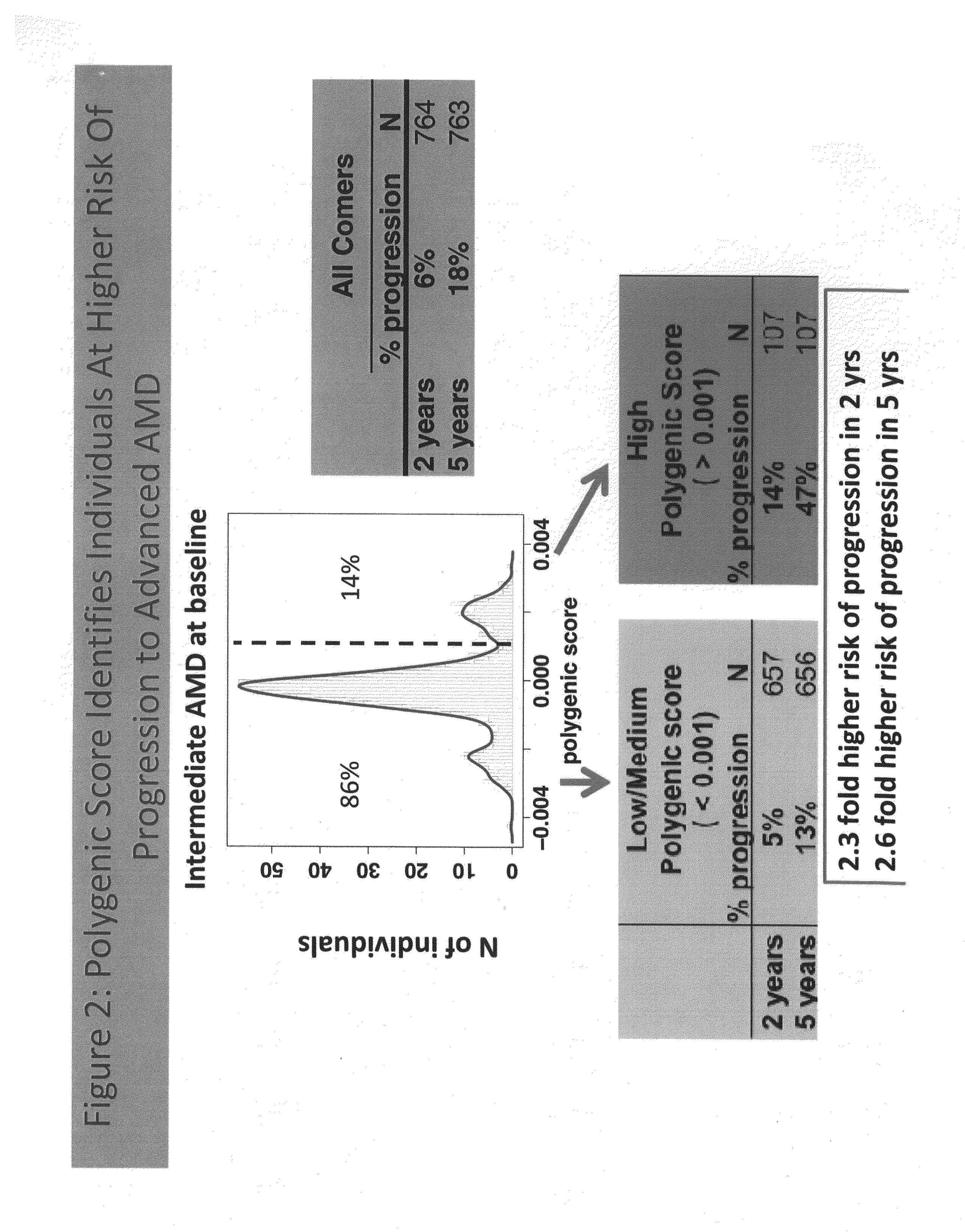
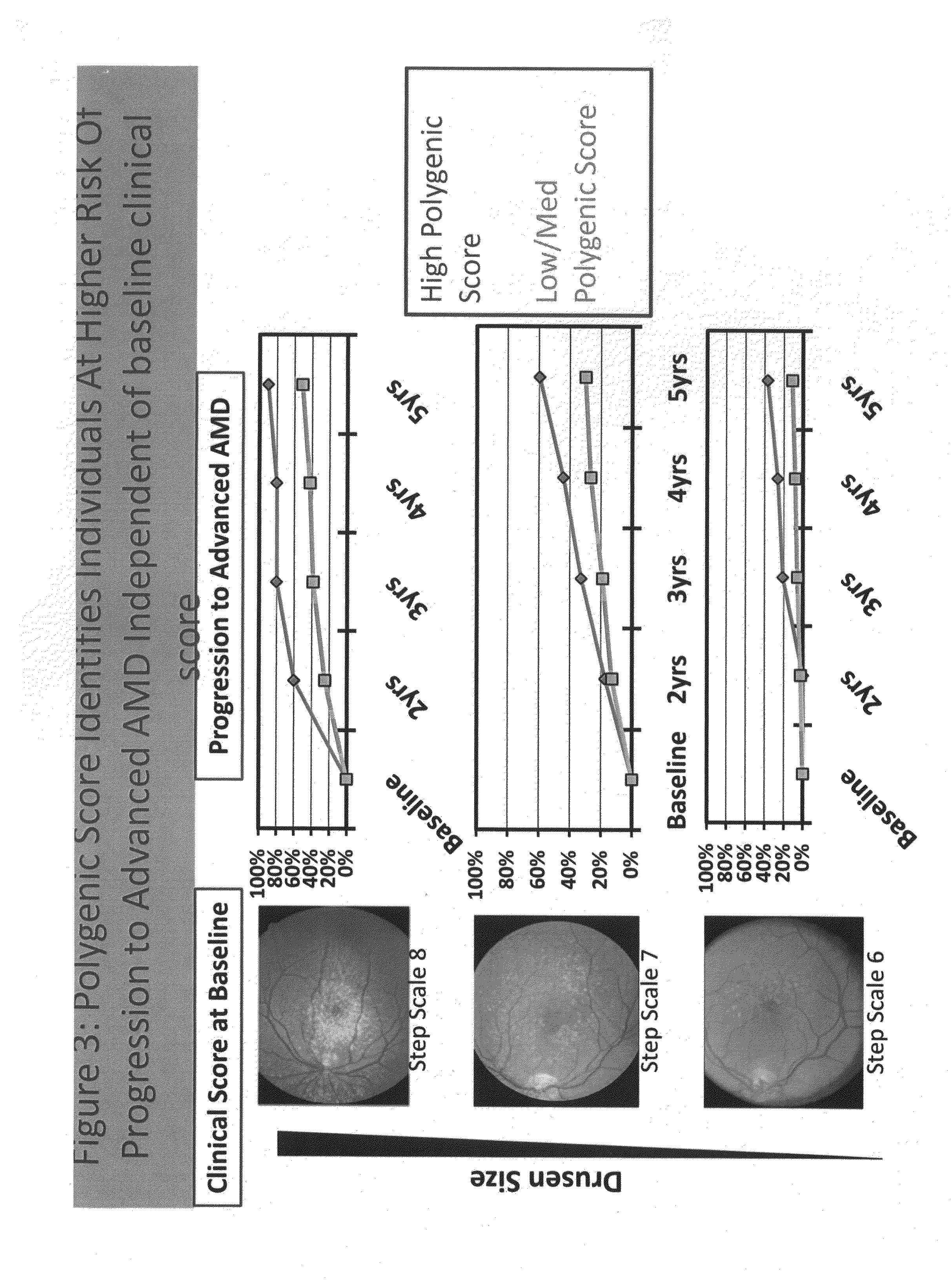
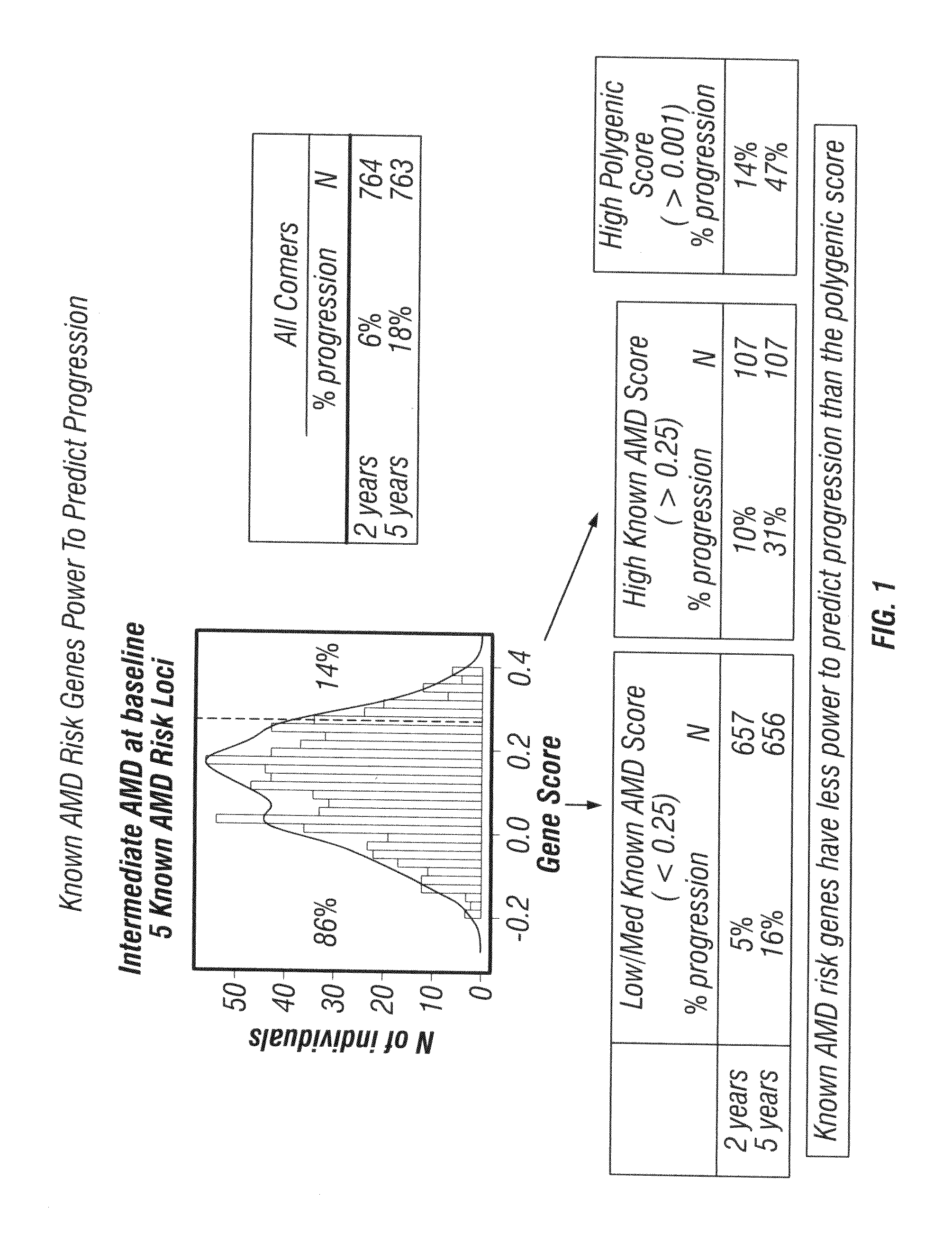
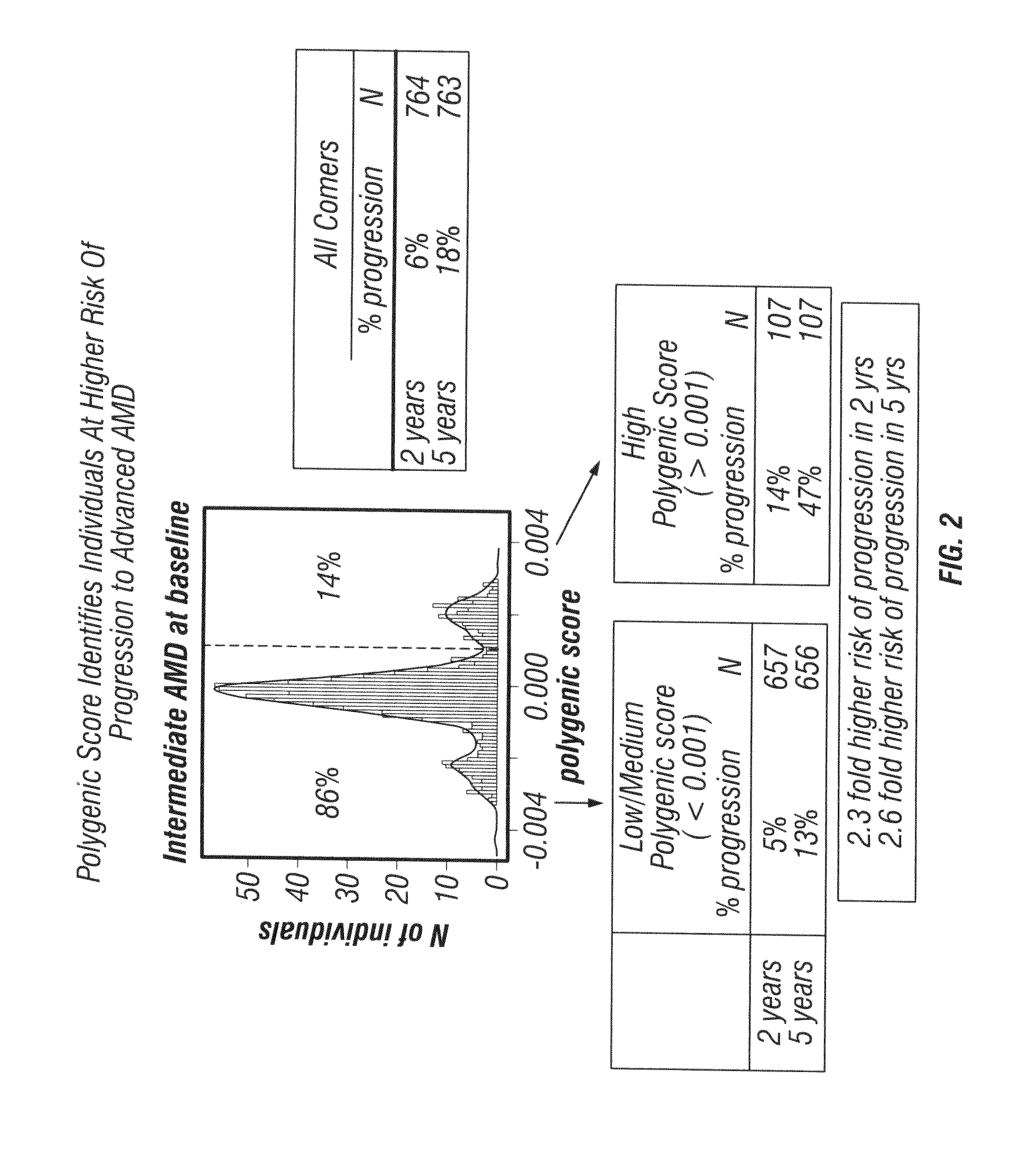
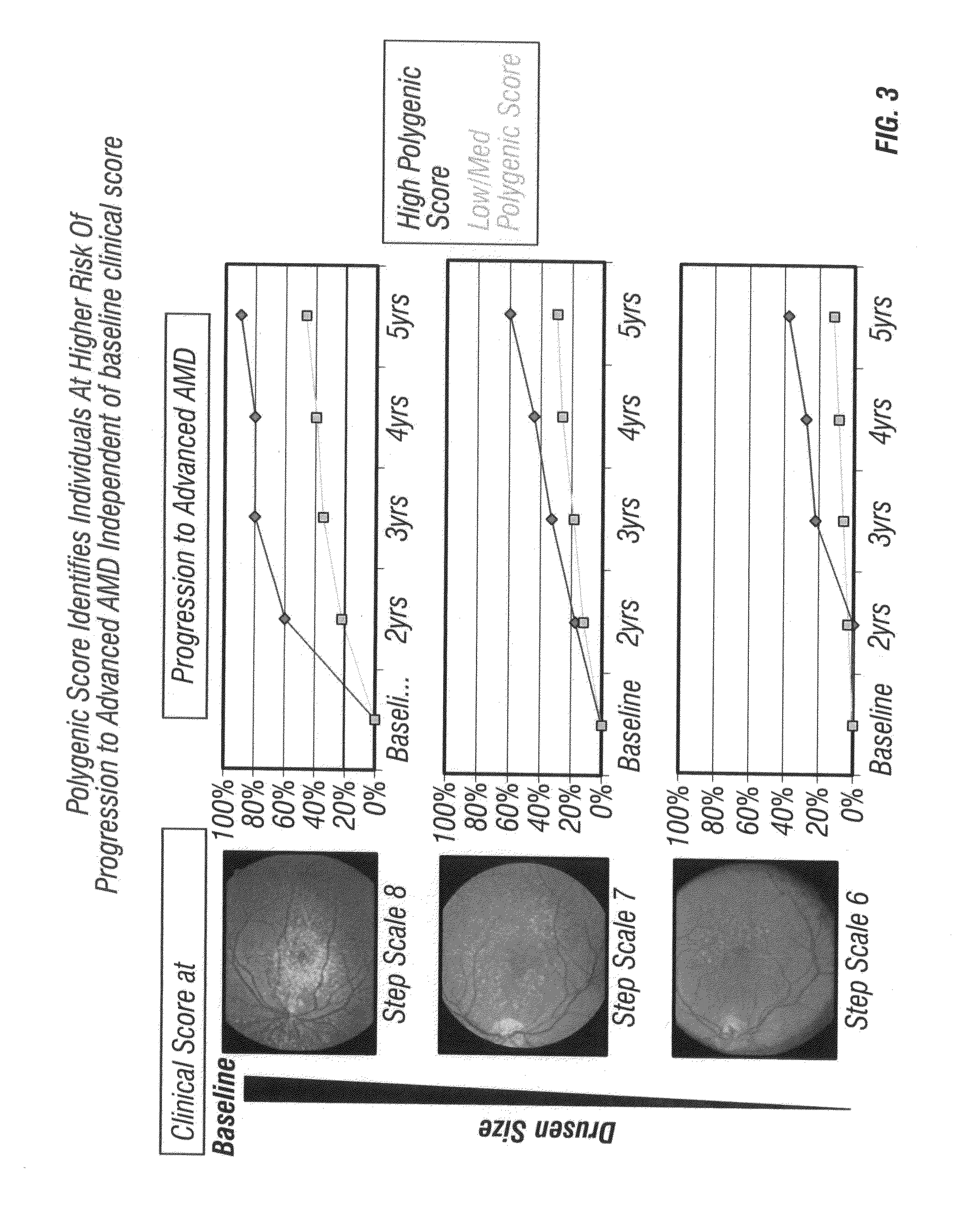
Predicting progression to advanced age-related macular degeneration using a polygenic score
The present invention relates to methods for identifying individuals with intermediate age-related macular degeneration (AMD) who possess a greater risk of progression to advanced AMD, using a polygenic score calculated based on the results of genome-wide gene association studies, using thousands of single-nucleotide polymorphisms (SNPs).
Owner:F HOFFMANN LA ROCHE & CO AG
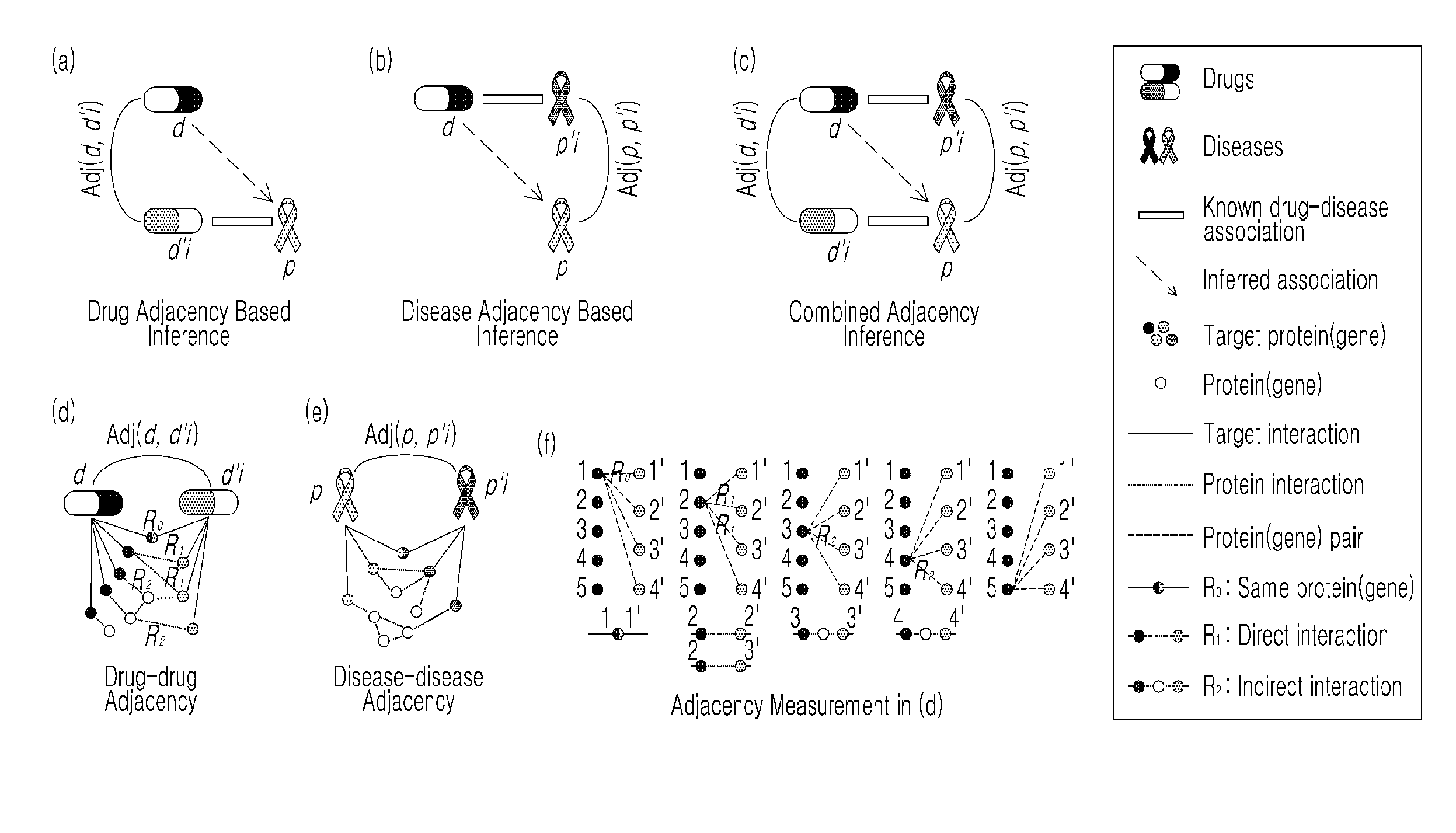
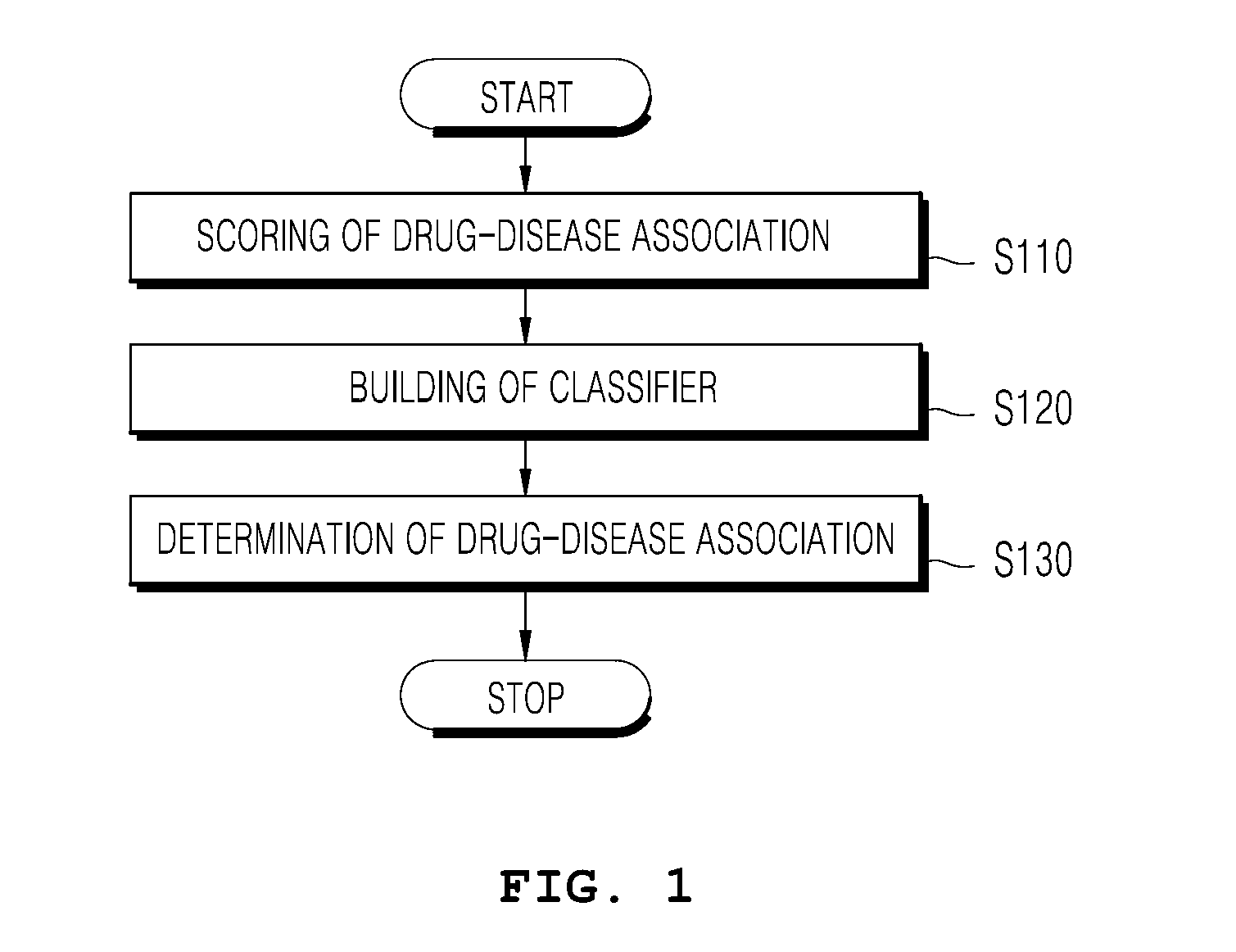
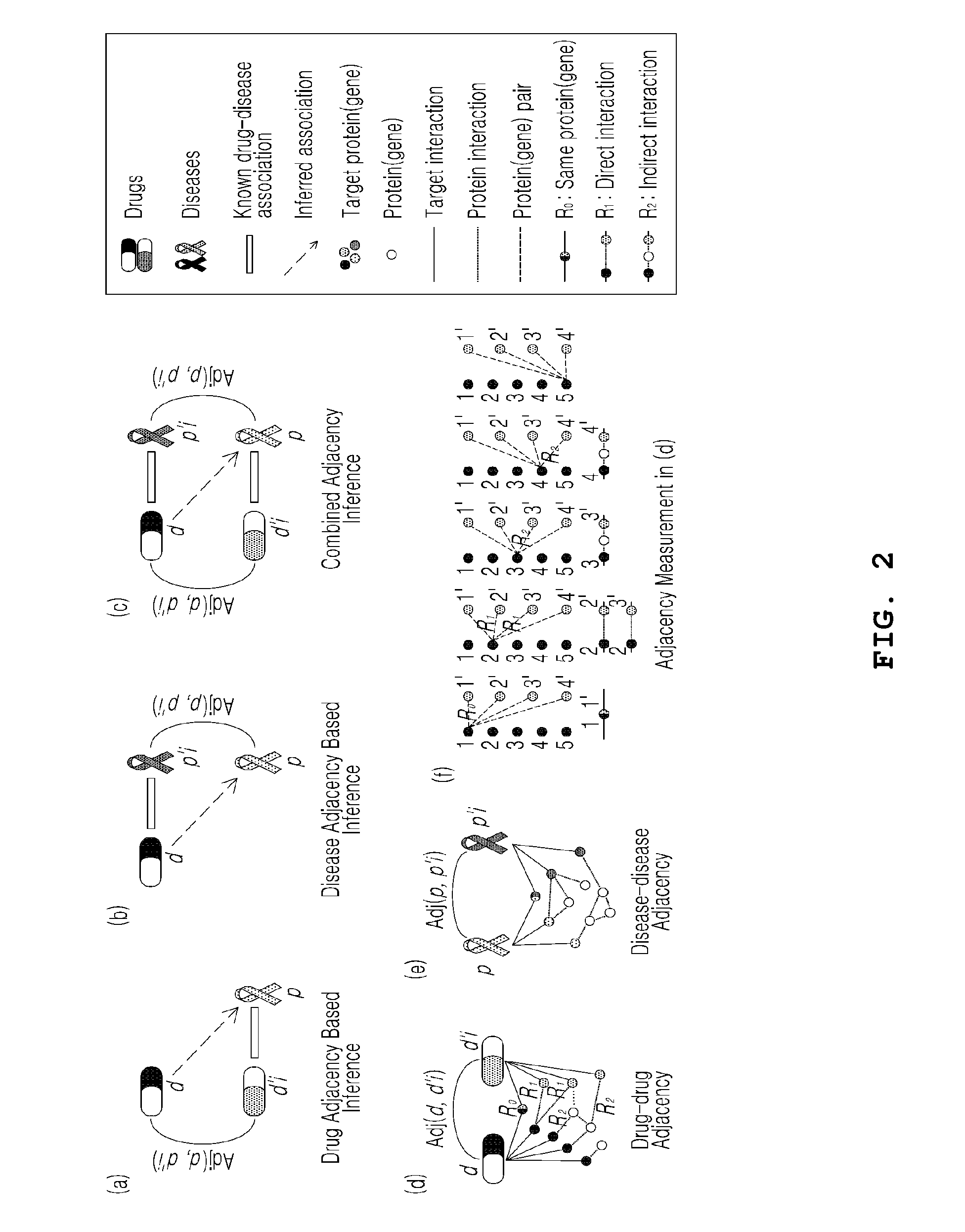
Apparatus and method for assessing effects of drugs based on networks
Disclosed is a network-based drug efficacy-assessing method, and an apparatus. The method comprises: computing a score for association between drugs and diseases by means of at least one of drug-adjacency-based inference, disease-adjacency-based inference, and module-distance-based inference; building a classifier for determining whether the drug is associated with the disease, using machine learning in which the score is employed as a feature; and determining the association between the drug and the disease by use of the classifier. The method and apparatus can search for drug-disease relation in which a drug can exert its pharmaceutical efficacy on a disease on the basis of a network constructed from protein interaction databases and protein-gene association databases, and can evaluate the molecular interaction of the drug to find out new pharmaceutical effects of the drug with higher precision and sensitivity, whereby a time and cost for the development of new drugs can be reduced.
Owner:GACHON UNIV OF IND ACADEMIC COOPERATION FOUND
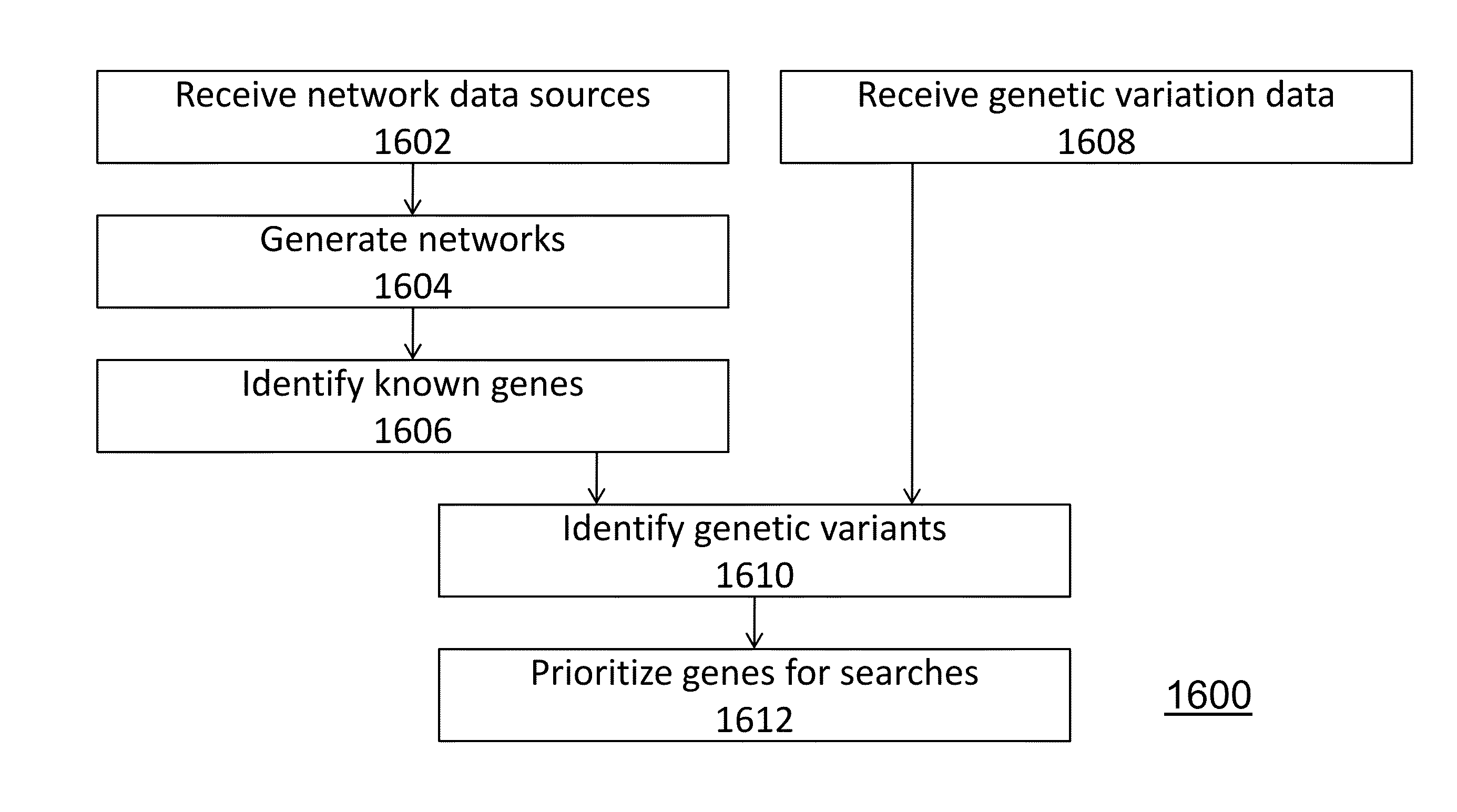
Method and system for network modeling to enlarge the search space of candidate genes for diseases
ActiveUS20130090908A1Analogue computers for chemical processesSystems biologyGenomic sequencingCandidate Gene Association Study
With the advent of low cost, high-throughput whole genome sequencing (“next generation sequencing”), tools are available to assay human genetic variation contributing to inherited disease syndromes. A method is disclosed for prioritization of genetic variants, and identification of disease genes, using network modeling of gene associations.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
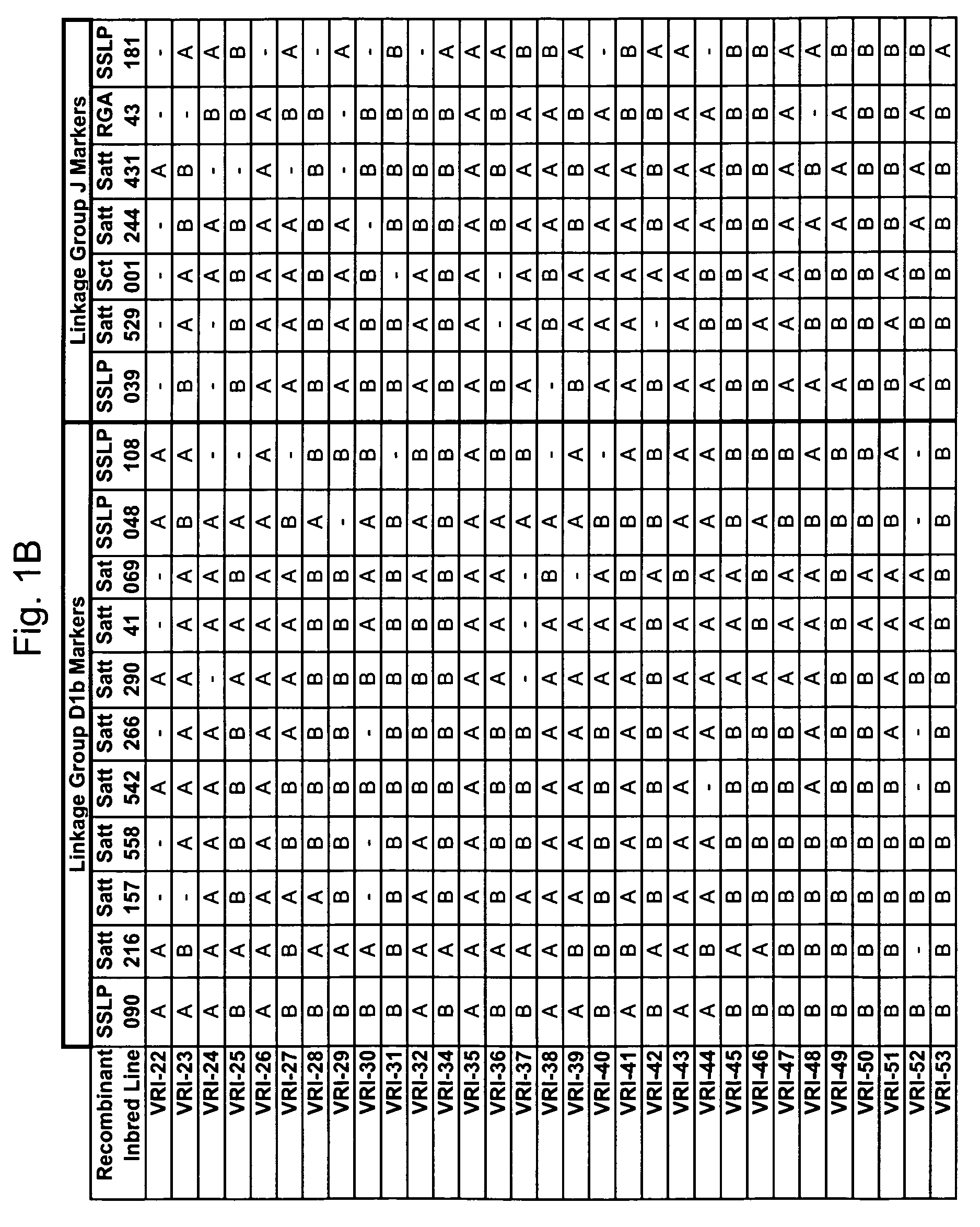
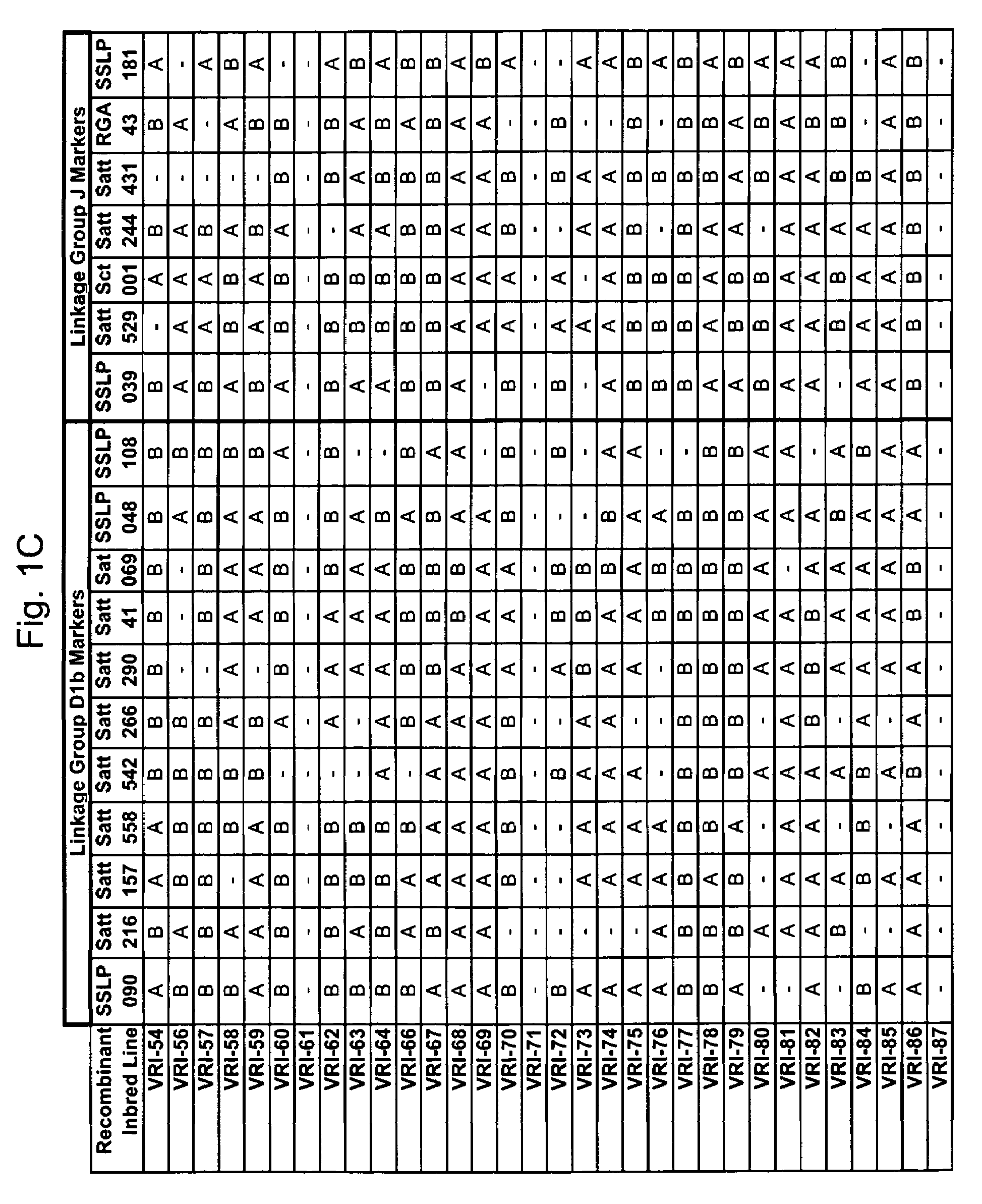
Marker mapping and resistance gene associations in soybean
InactiveUS7642403B2Microbiological testing/measurementOther foreign material introduction processesMarker-assisted selectionResistant genes
The invention provides novel molecular genetic markers in soybean, where the markers are useful, for example, in the marker-assisted selection of gene alleles that impart disease-resistance, thereby allowing the identification and selection of a disease-resistant plant. The markers also find use in positional cloning of disease-resistance genes.
Owner:VIRGINIA TECH INTPROP INC +1
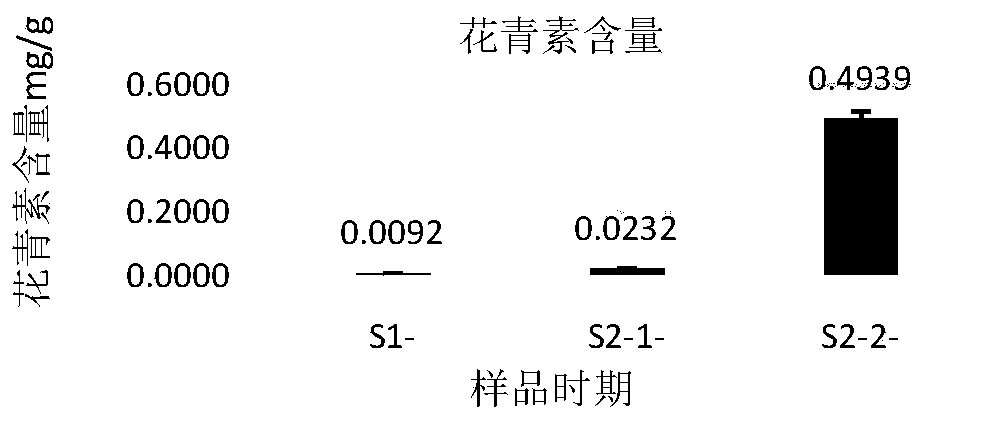
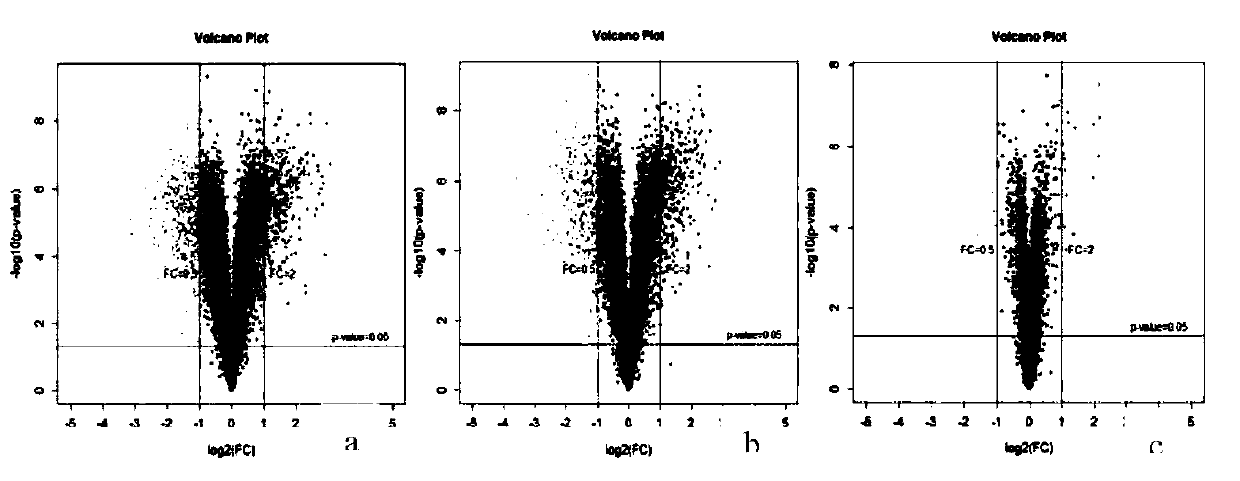
Analysis method for researching formation of variegation of nelumbo nucifera based on combination of transcriptomics with proteomics TMT
PendingCN110331225ADecor optimizationQuantitative and qualitativeMicrobiological testing/measurementBiological material analysisBiologyGene association
The invention relates to an analysis method for researching formation of variegation of nelumbo nucifera based on combination of transcriptomics with proteomics TMT, and provides a new method for disclosing a molecular mechanism of the formation of the variegation of plants. Samples of "Dasajin" nelumbo nucifera petals are collected, through transcriptome Illumina HiSeq sequencing, expression difference genes on an anthocyan metabolic pathway are screened, besides, proteins in the samples are extracted, the concentration of the proteins is determined, the quality is detected, through TMT labelmarking and 2D-LC-MS analysis, bioinformatics analysis is performed through combining with the proteomics, the proteins for adjusting and controlling expressing level differences on the anthocyan metabolic pathway are screened, the genes having differences in the transcriptome are related into the proteomics, and key proteins and corresponding genes for adjusting and controlling the variegation of "Dasajin" nelumbo nucifera are screened, so that the purpose of understanding a manner of the proteins and the genes for adjusting the formation of the variegation of the "Dasajin" nelumbo nuciferais achieved, and a new thinking and a new method are provided for the formation mechanism of the variegation of other plants.
Owner:CHINA THREE GORGES CORPORATION
Pathogenic microorganism virulence gene association model as well as establishment method and application thereof
PendingCN112837745AAccurately determine the pathogenicitySampling needs are smallProteomicsGenomicsGenomic sequencingPathogenic microorganism
The invention relates to a pathogenic microorganism virulence gene association model and an establishment method and application thereof, and belongs to the technical field of gene detection. The method comprises the following steps: establishing a pathogenic microorganism gene database and a virulence gene database; acquiring pathogenic microorganism metagenome sequencing data of clinical samples to obtain pathogenic microorganisms in each clinical sample and corresponding virulence gene sequence data; carrying out clustering analysis on the sequence data of the pathogenic microorganisms and the sequence data of the virulence genes to obtain a normal distribution model of a single virulence gene and at least one suspected associated pathogenic microorganism, and selecting a model with high pathogenic microorganism abundance and strong correlation between the virulence gene sequence number and the pathogenic microorganism sequence number. The pathogenic microorganism virulence gene correlation model obtained by the method can judge which bacteria each virulence gene comes from, so that the model overcomes the defects of the prior art, and has the advantages of wide application range and high accuracy.
Owner:GZ VISION GENE TECH CO LTD +3
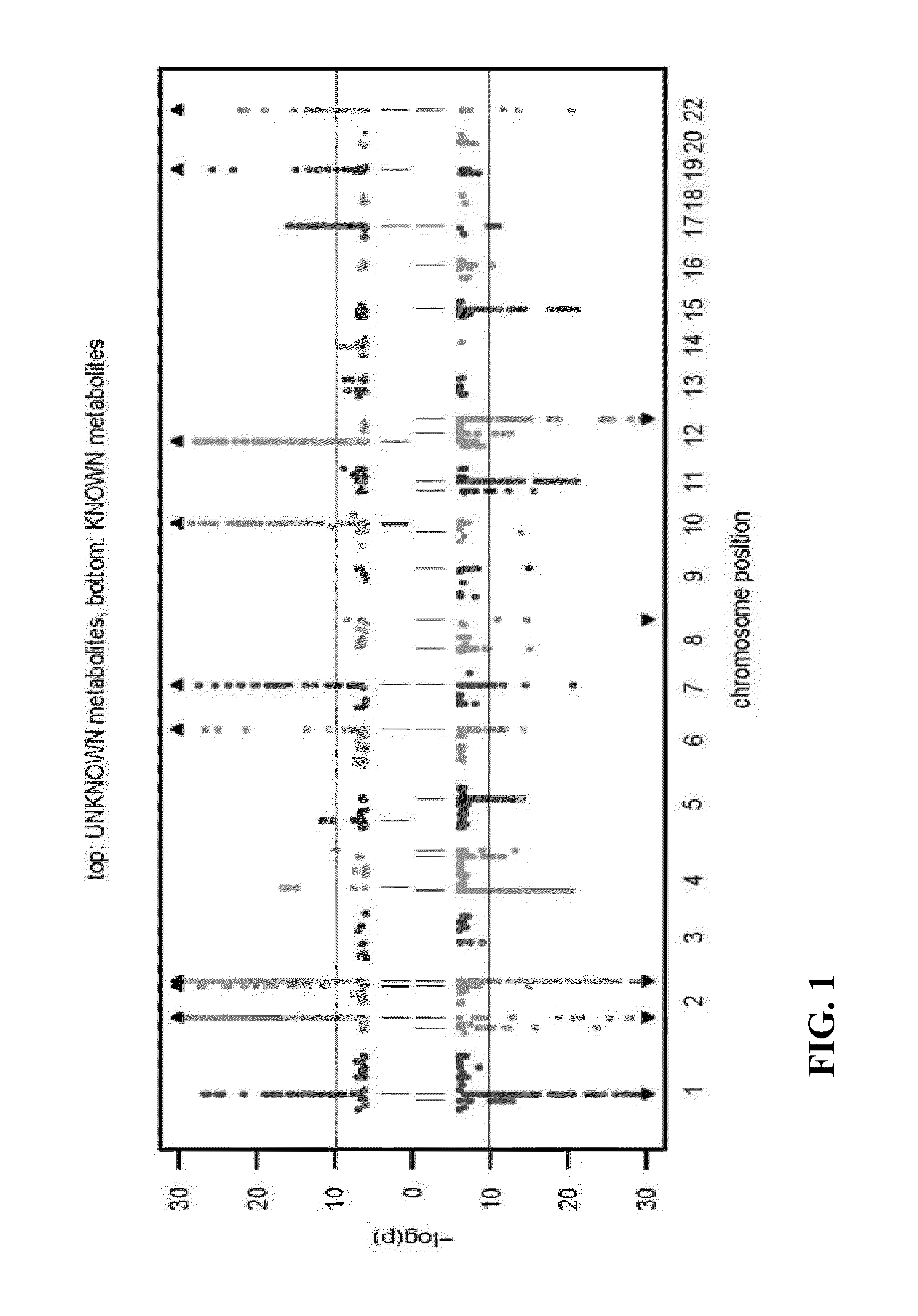
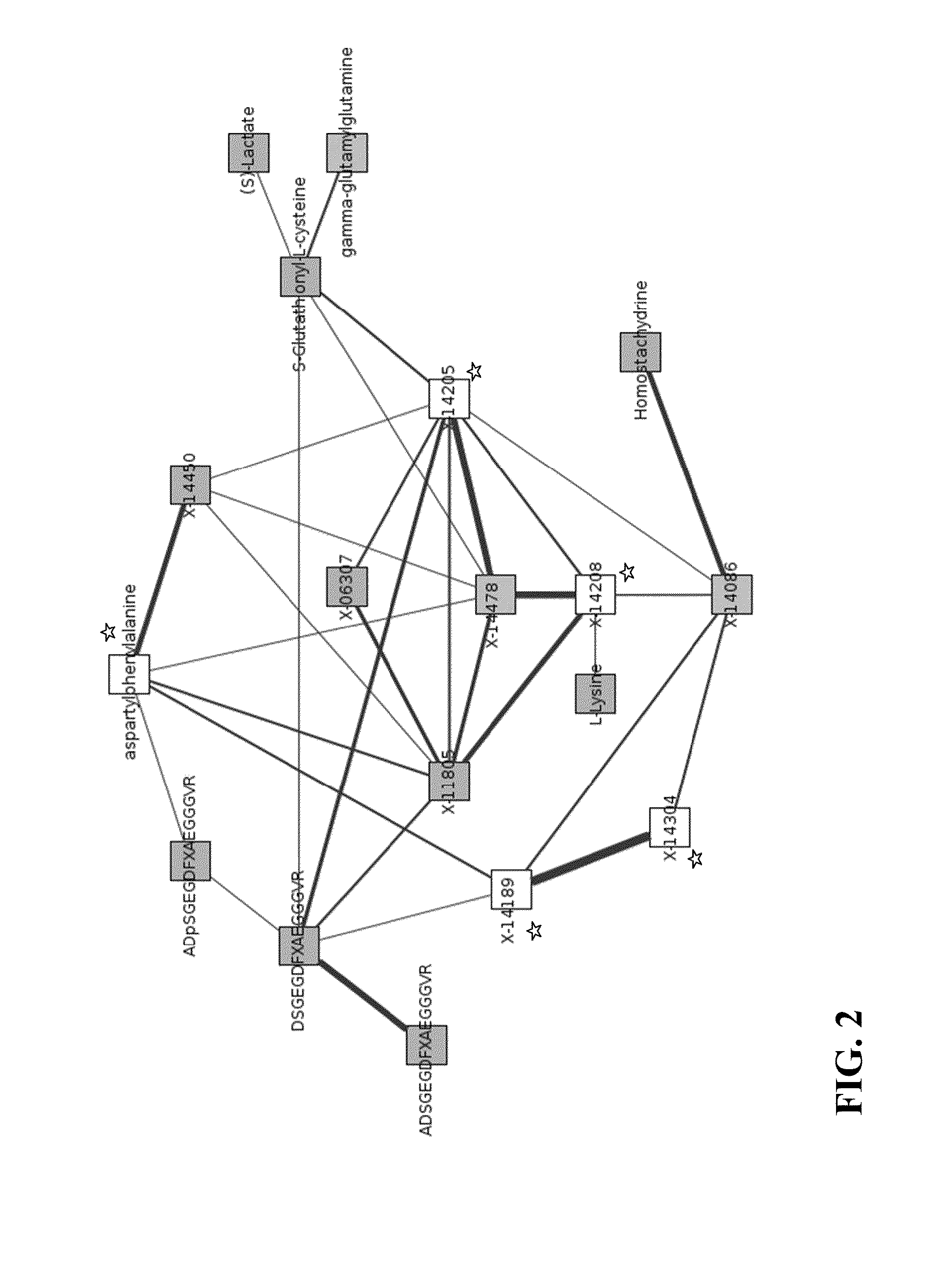
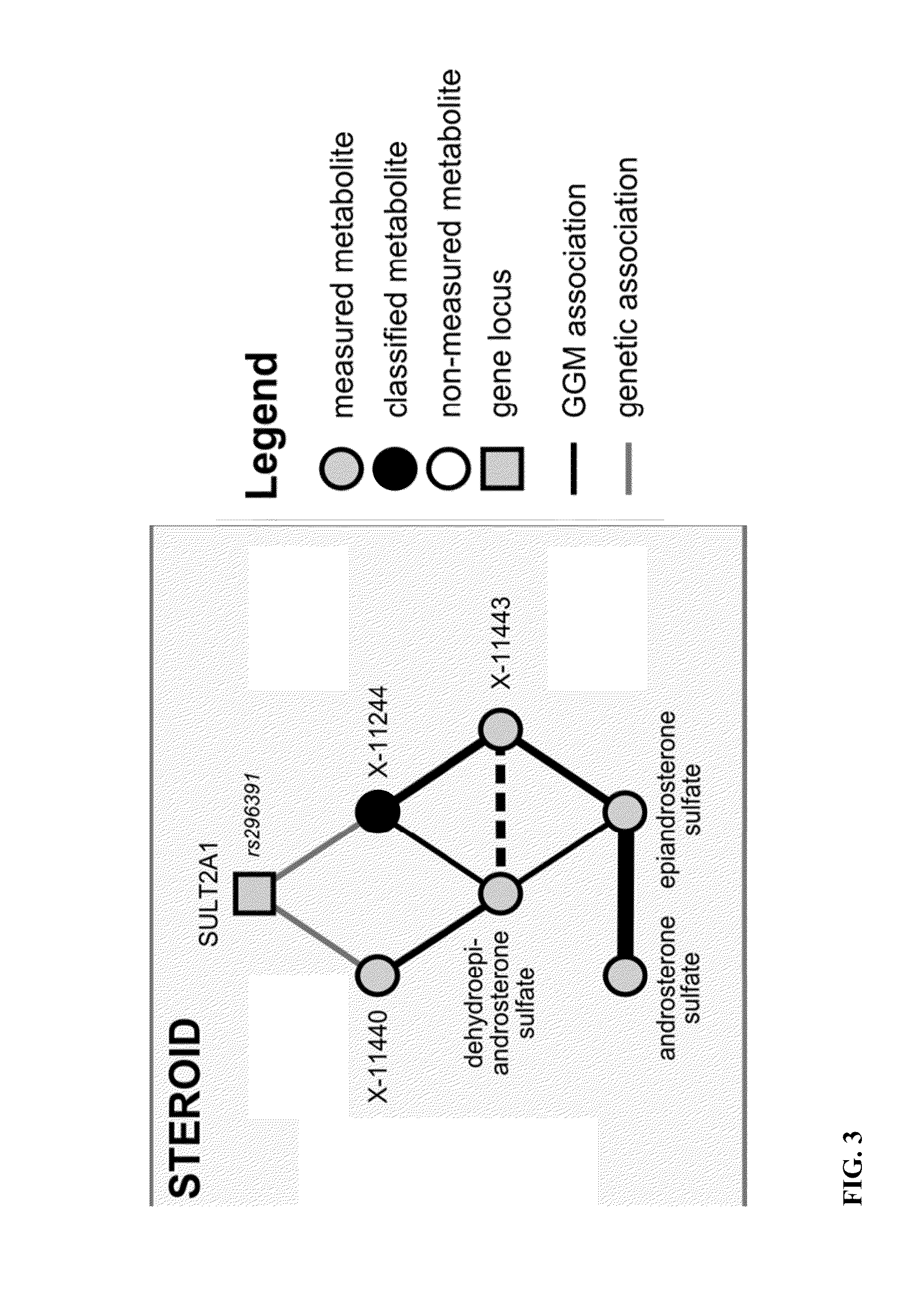
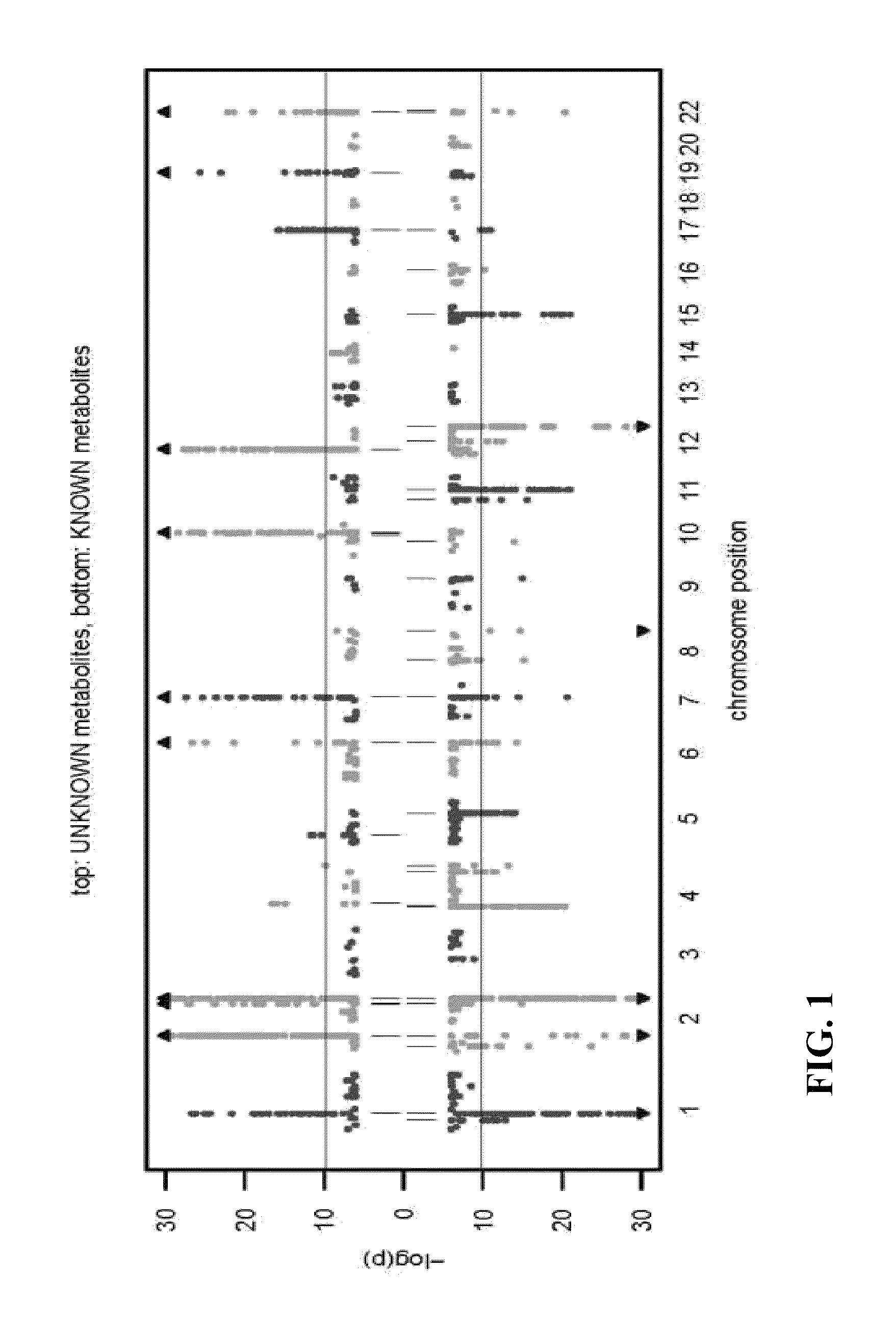
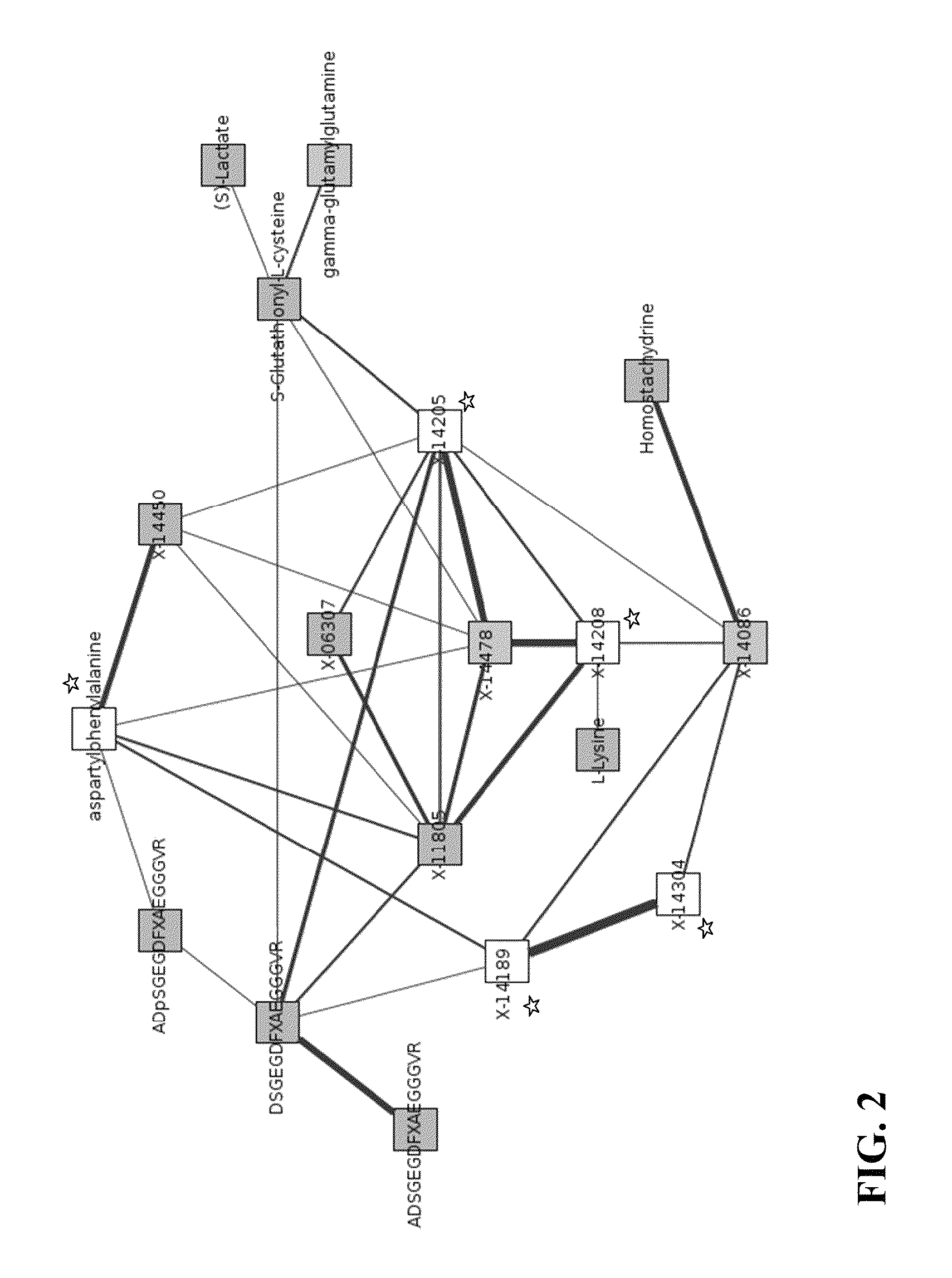
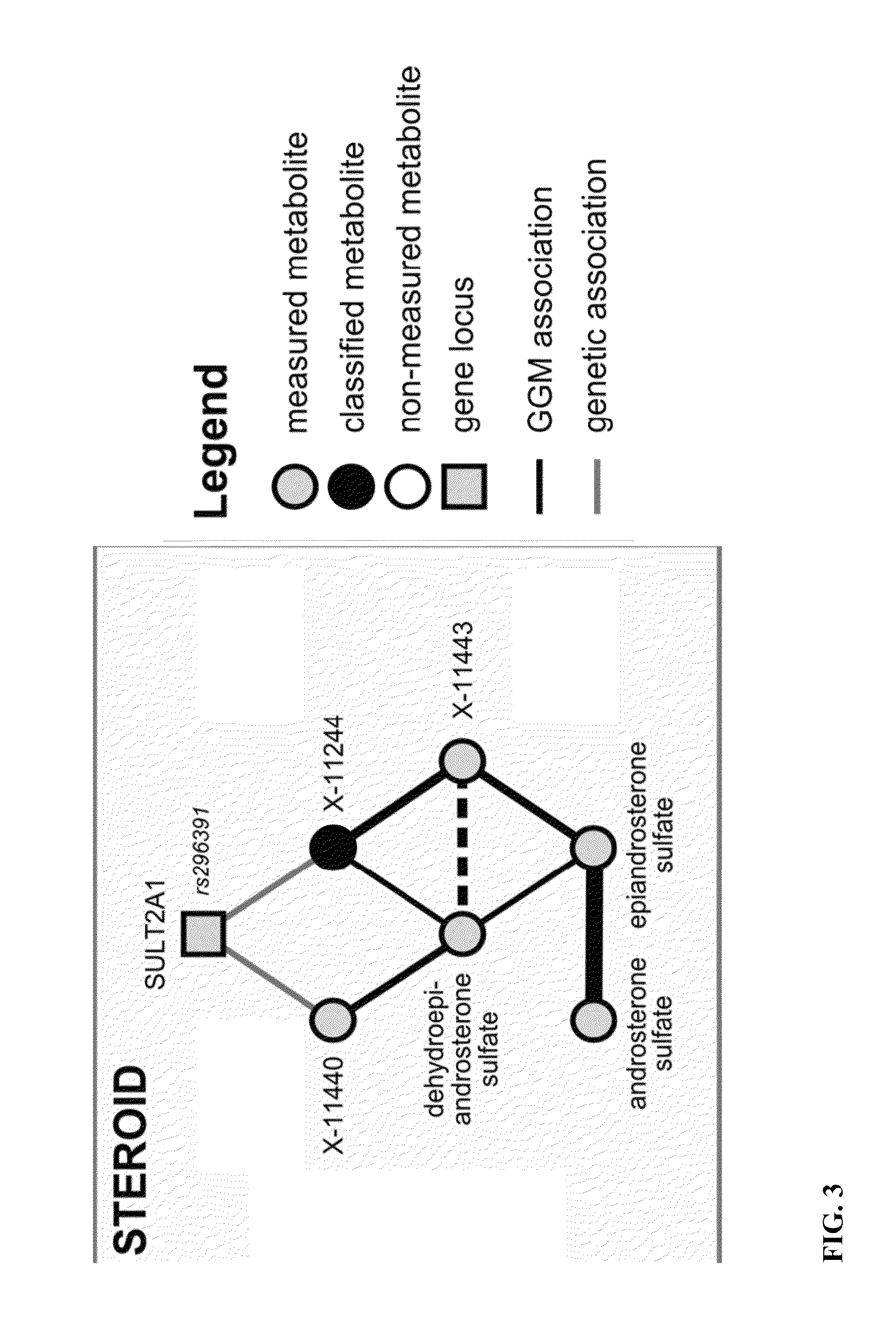
Identity Elucidation of Unknown Metabolites
A method of elucidating the identity of an unknown metabolite comprising measuring amounts of known and unknown metabolites in subjects; associating the unknown metabolite with a specific gene from a gene association study; determining a protein associated with the specific gene and analyzing information for the protein; associating the unknown metabolite with concentrations and / or ratios of other metabolites in subjects; obtaining chemical structural data for the unknown metabolite; and using the information obtained to elucidate the identity of the unknown metabolite.
Owner:METABOLON +1
Predicting progression to advanced age-related macular degeneration using a polygenic score
The present invention relates to methods for identifying individuals with intermediate age-related macular degeneration (AMD) who possess a greater risk of progression to advanced AMD, using a polygenic score calculated based on the results of genome-wide gene association studies, using thousands of single-nucleotide polymorphisms (SNPs).
Owner:GENENTECH INC
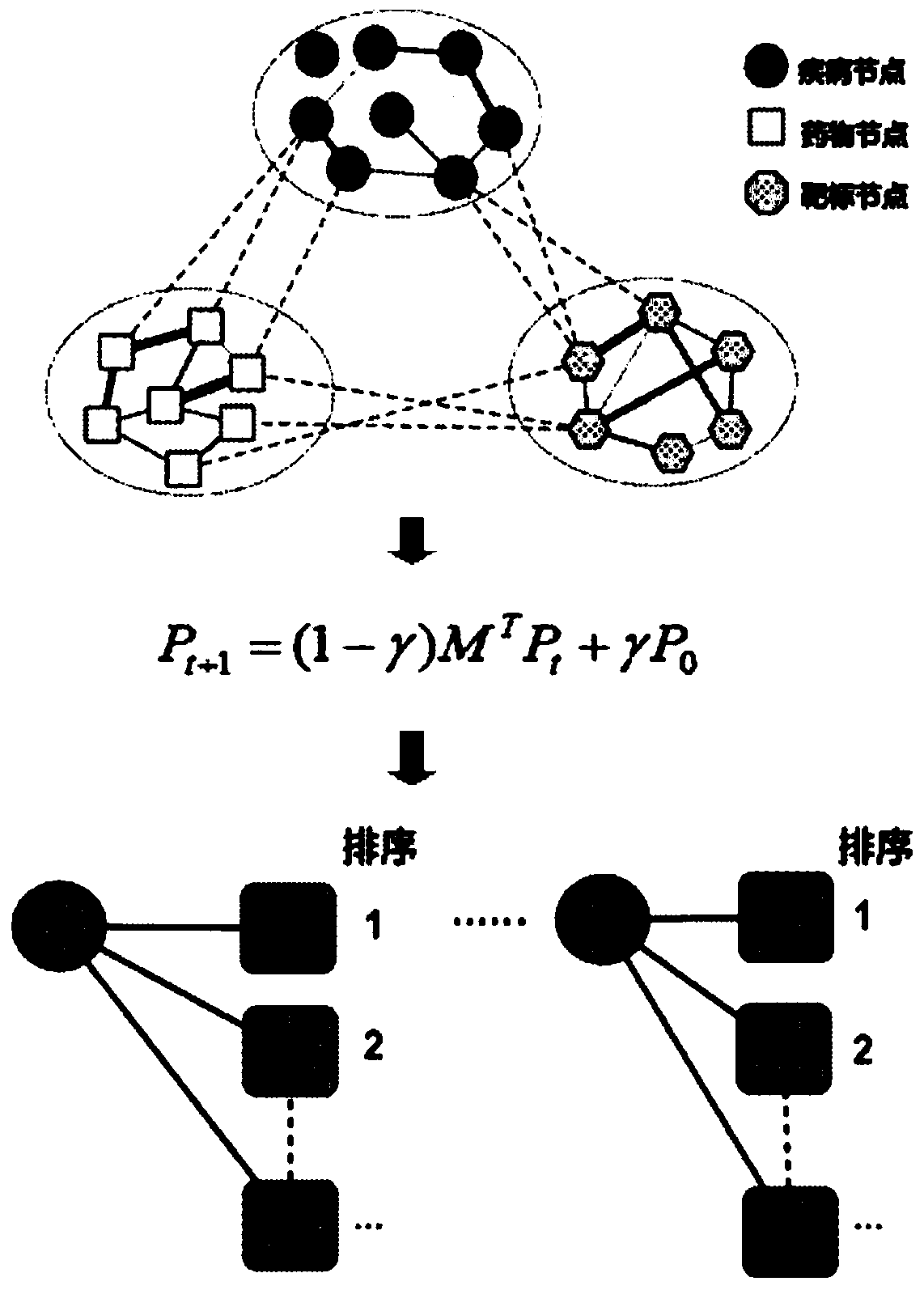
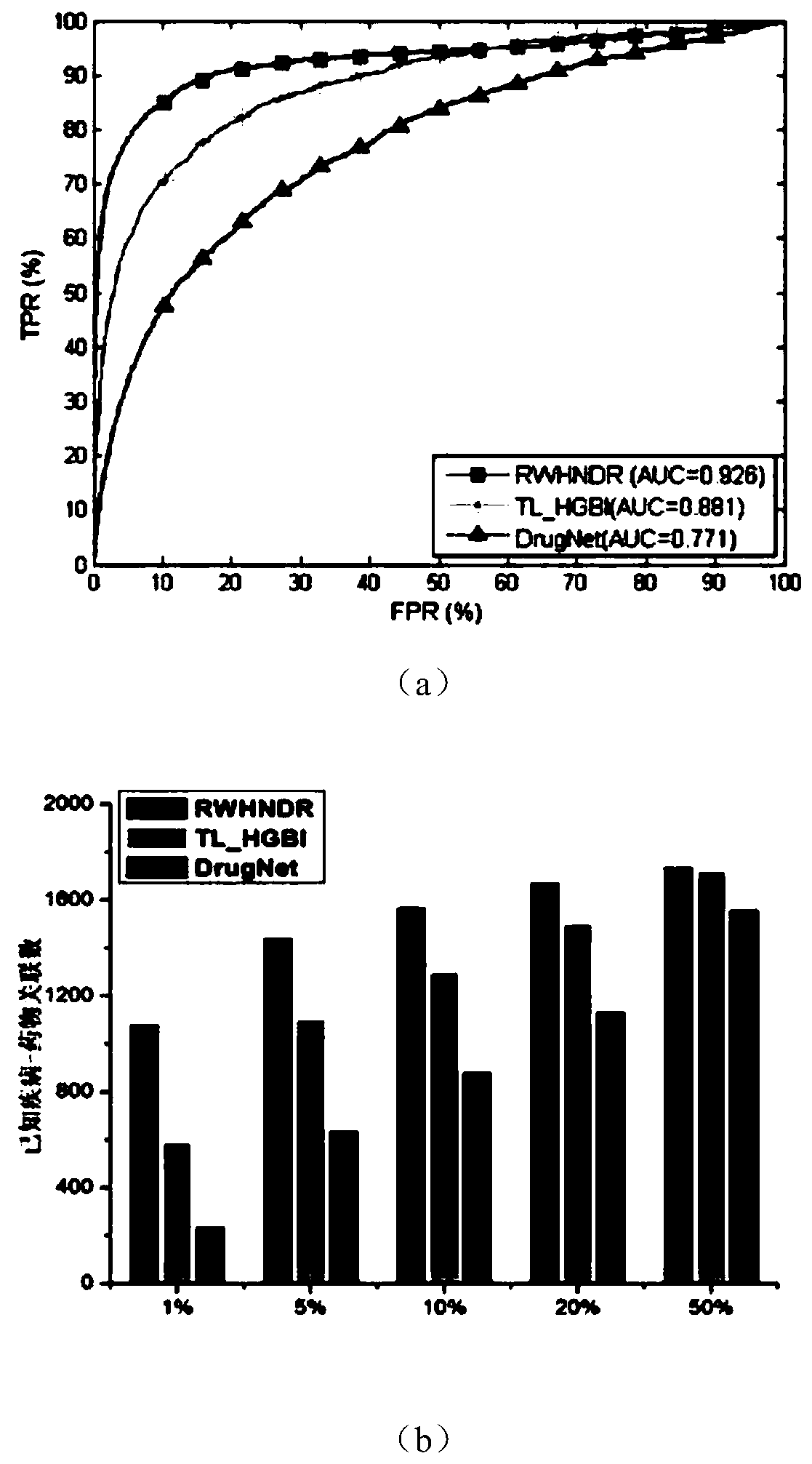
A drug repositioning method based on multivariate information fusion and random walk model
ActiveCN107506591BEfficient miningTake advantage ofMolecular designDrug referencesData setHeterogeneous network
The invention discloses a drug repositioning method based on multi-information fusion and a random walk model. According to the method, disease-target-drug heterogeneous network is constructed through integrating existing disease data, drug data, target data, disease-drug associated data, disease-gene associated data and drug-target associated data; the basic random walk model is extended to the constructed heterogeneous network; and candidate therapeutic drugs are recommended for diseases through effectively utilizing global network information. The method disclosed by the invention is simple and effective; and compared with other methods and proved by tests on a standard data set, the method has good prediction performance in the aspect of drug repositioning.
Owner:CENT SOUTH UNIV
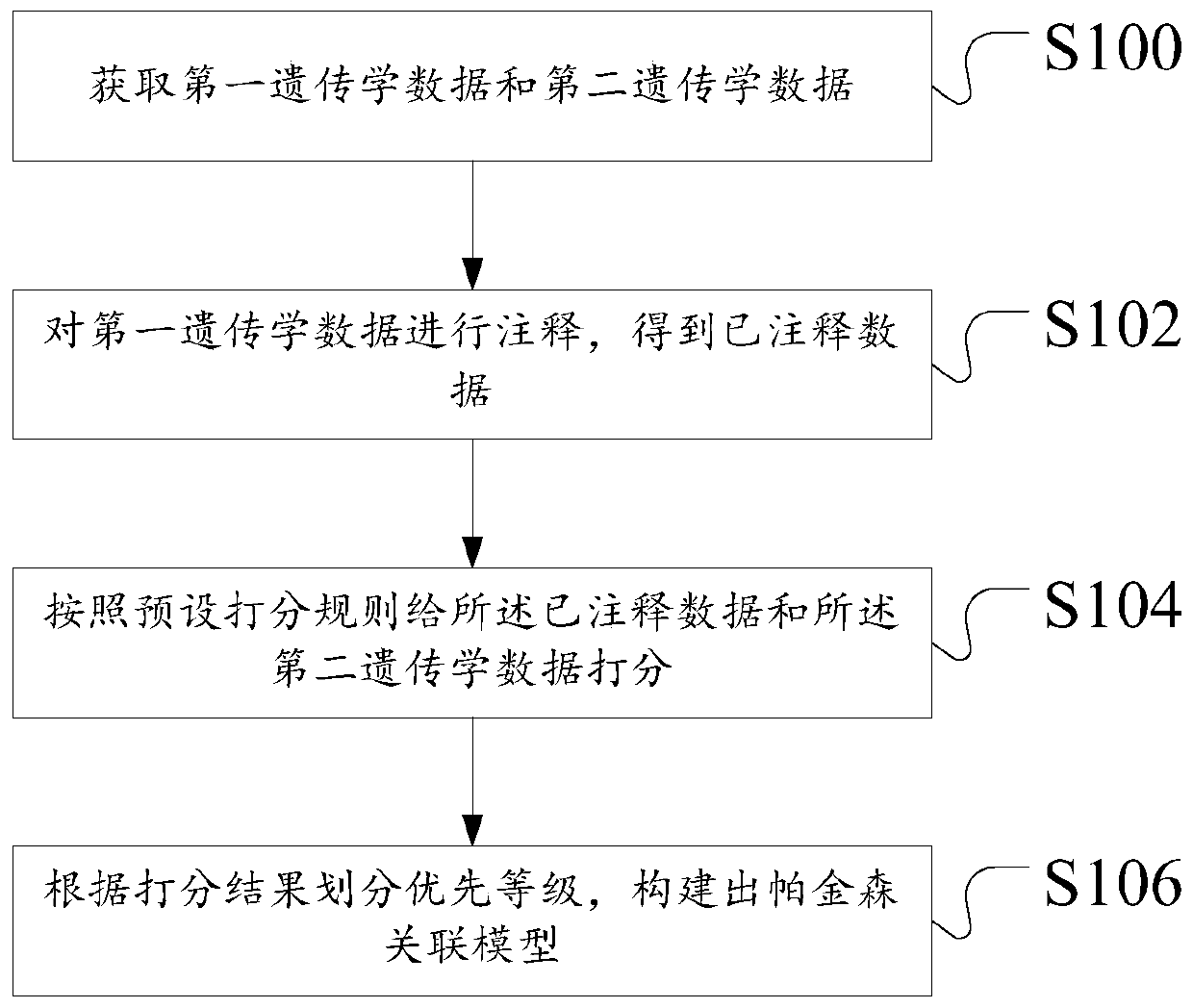
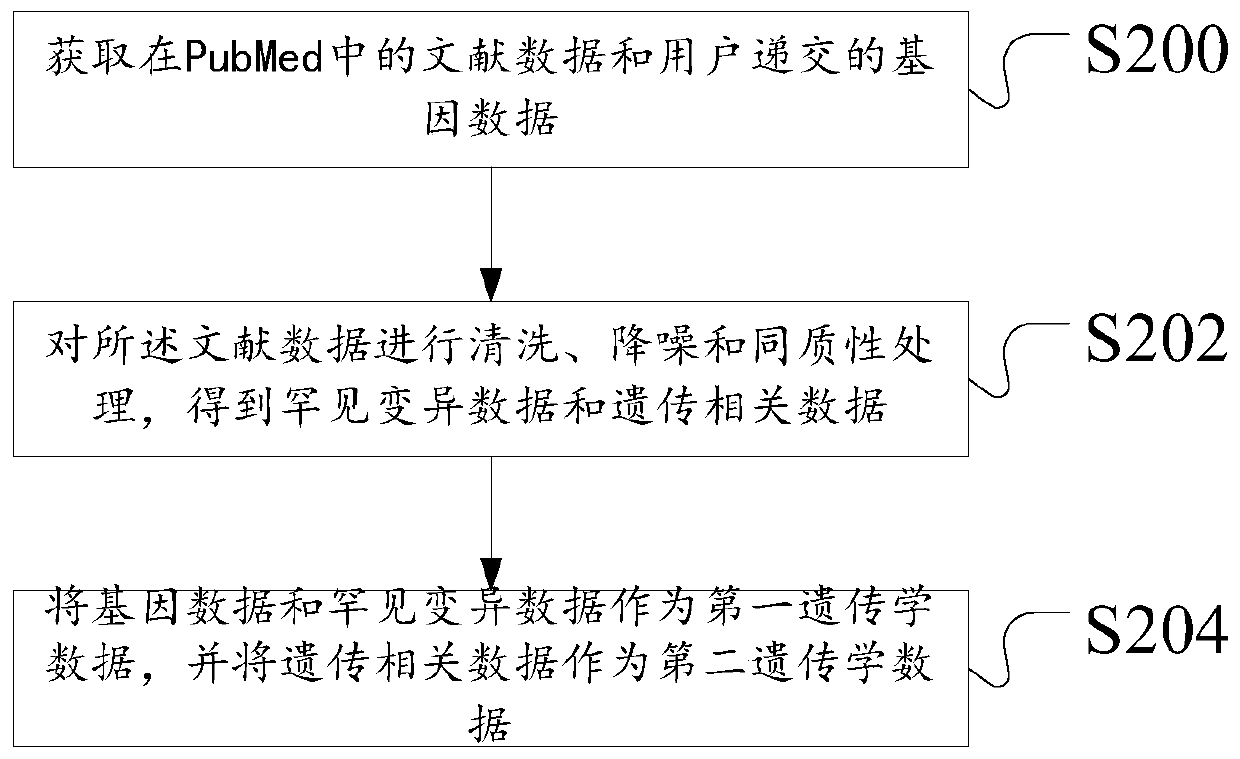
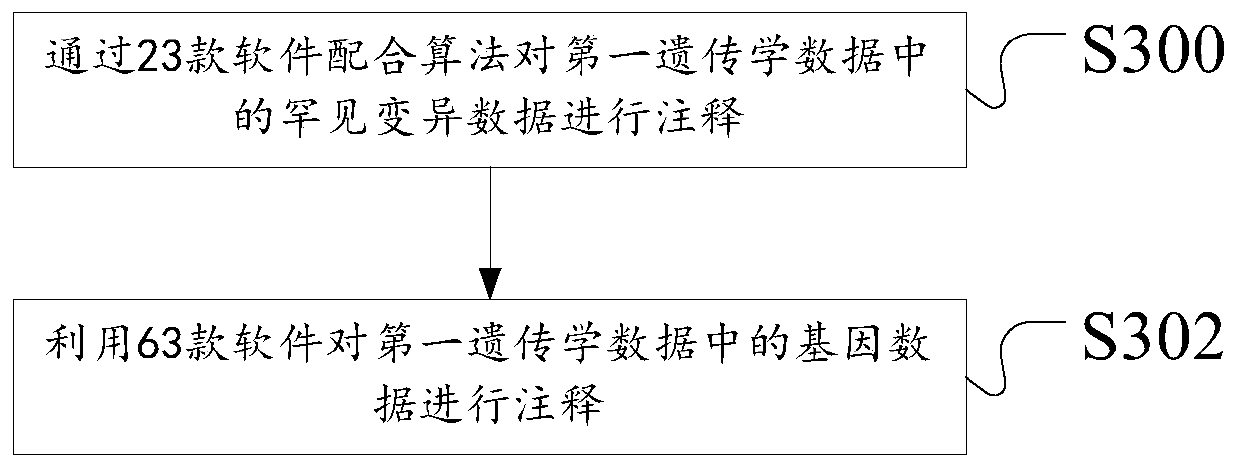
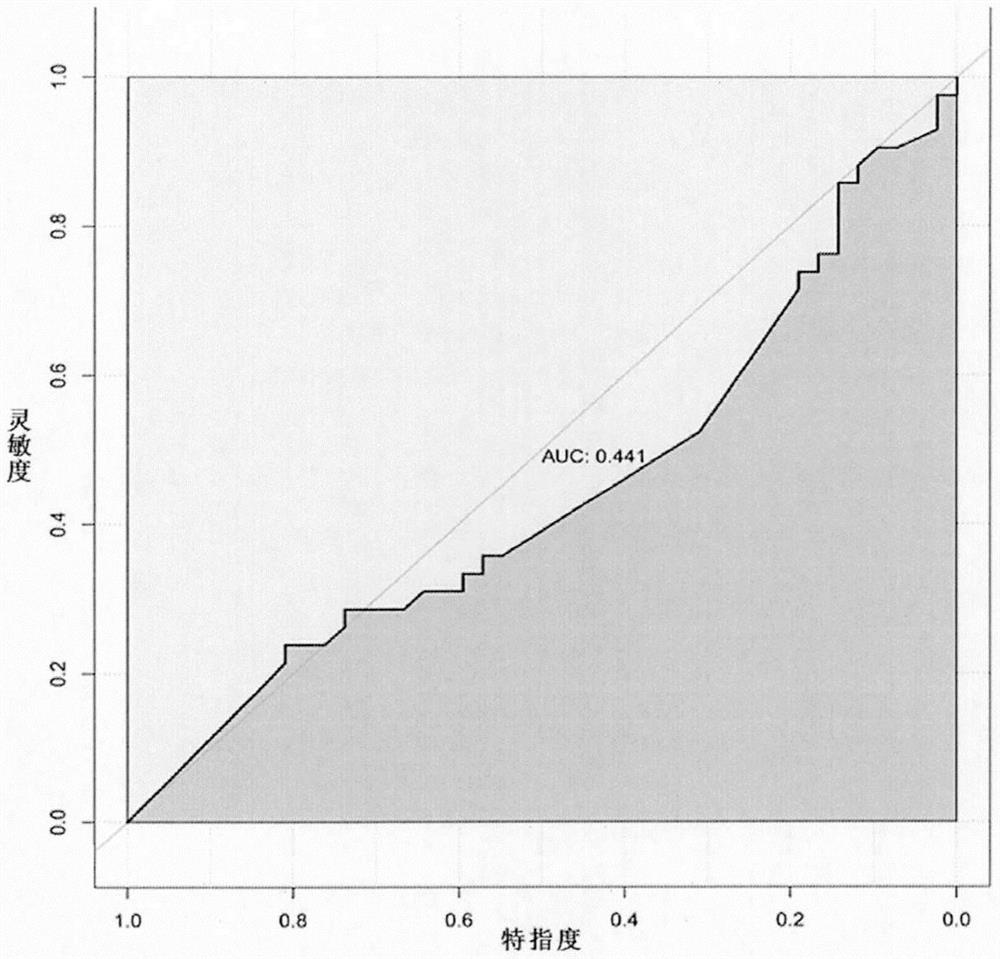
Construction method and device of Parkinson's disease genomics association model, server and storage medium
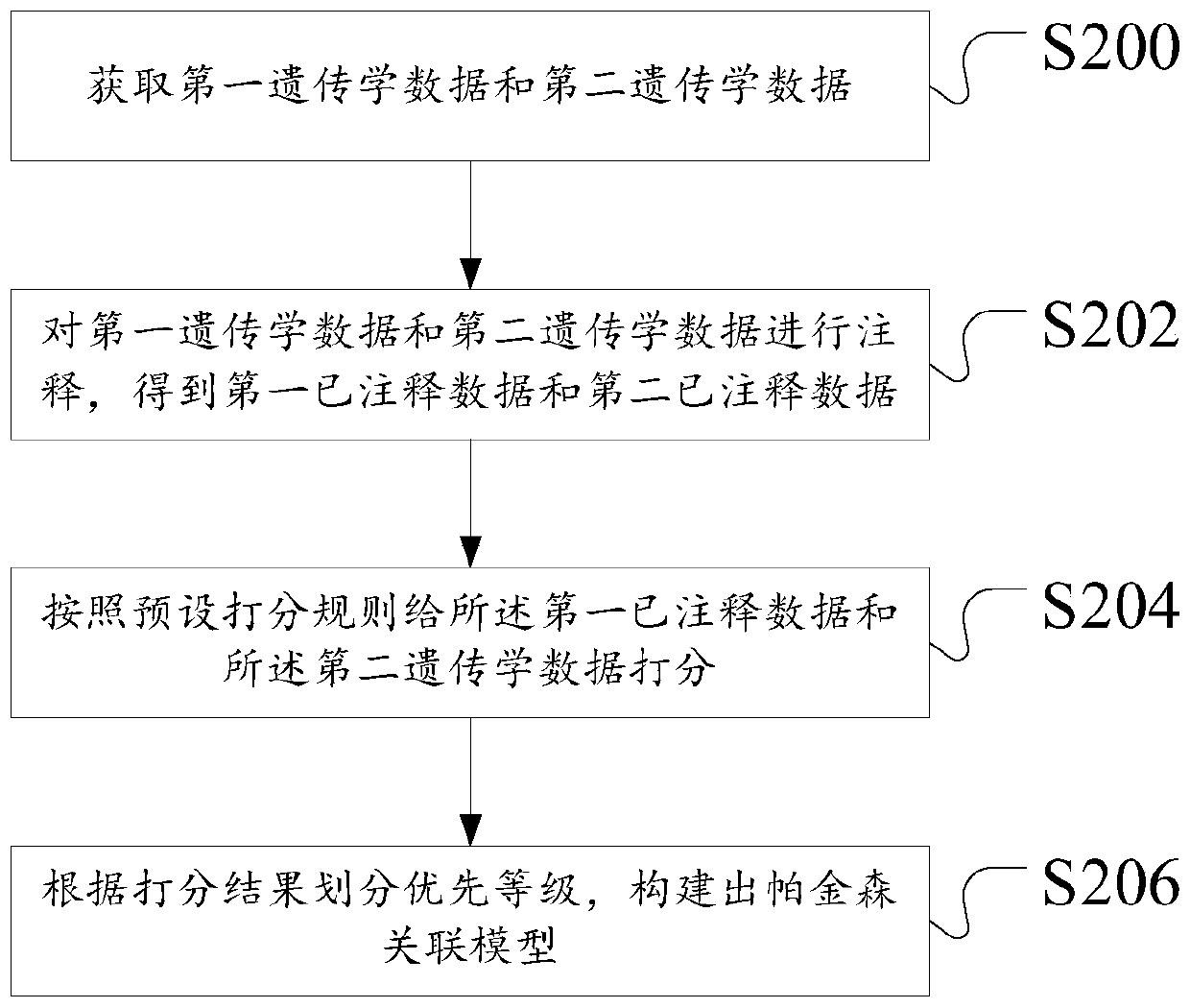
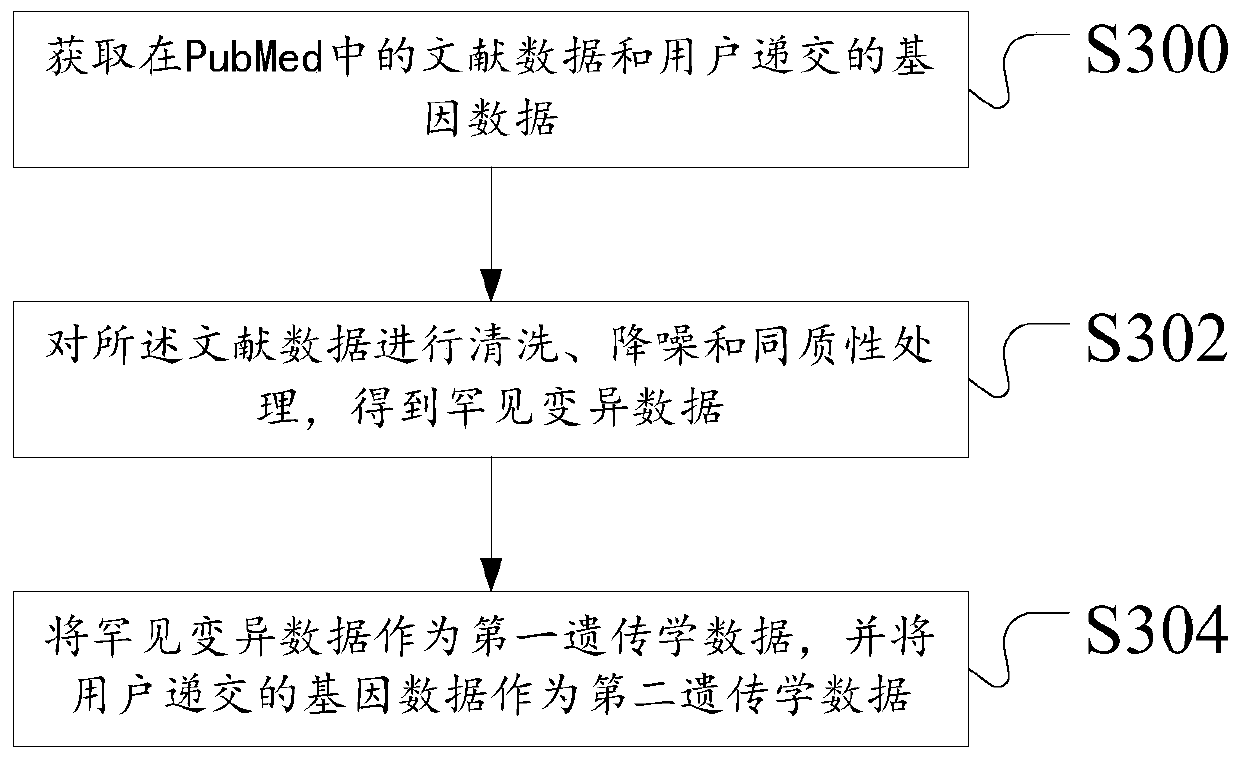
ActiveCN111192626ASimple Association JudgmentQuickly realize association judgmentBiostatisticsProteomicsGenomicsAssociation model
The invention discloses a construction method of a Parkinson's disease genomics association model. The method comprises the following steps: acquiring first genetics data and second genetics data; annotating the first genetics data to obtain annotated data; scoring the annotated data and the second genetics data according to a preset scoring rule; and dividing priority levels according to a scoring result, and constructing a Parkinson's correlation model. The technical problem that the Parkinson's disease and gene association judgment cannot be simply and quickly realized is solved.
Owner:XIANGYA HOSPITAL CENT SOUTH UNIV
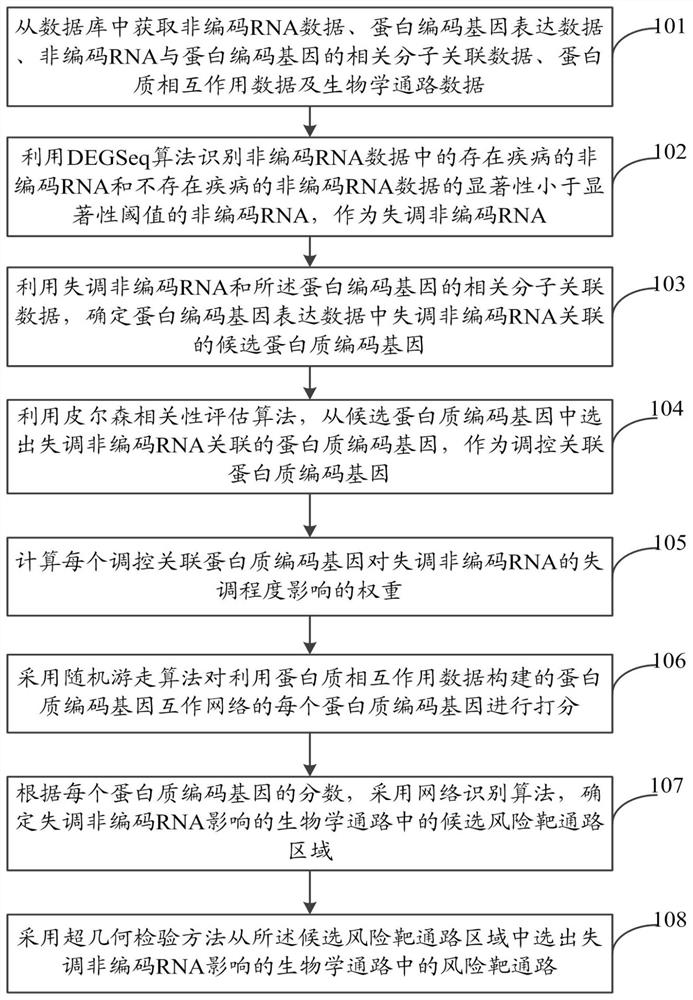
Method and system for identifying non-coding RNA (lnRNA) regulated disease risk target pathways
The invention discloses a method and a system for identifying lnRNA regulated disease risk target pathways. According to the method, risk pathway regions regulated and controlled by lncRNA in different disease types can be finely identified based on a lncRNA expression profile, an lncRNA-protein coding gene association interaction network and the expression disorder degrees of genes, which are located in pathways, in diseases, and taking multiple factors such as the association of the genes in the pathways and expression disorder lncRNA, the expression disorder degrees of the genes and the topological structures of the pathways into consideration; and the risk target pathways influenced by lncRNA regulation and control in complex diseases can be systematically identified, so the regulationand control effect and the functions of lncRNA in diseases can be revealed, and a novel reference is provided for the research of the pathogenesis of complex diseases from the perspective of non-coding RNA regulation and control.
Owner:李霞 +2
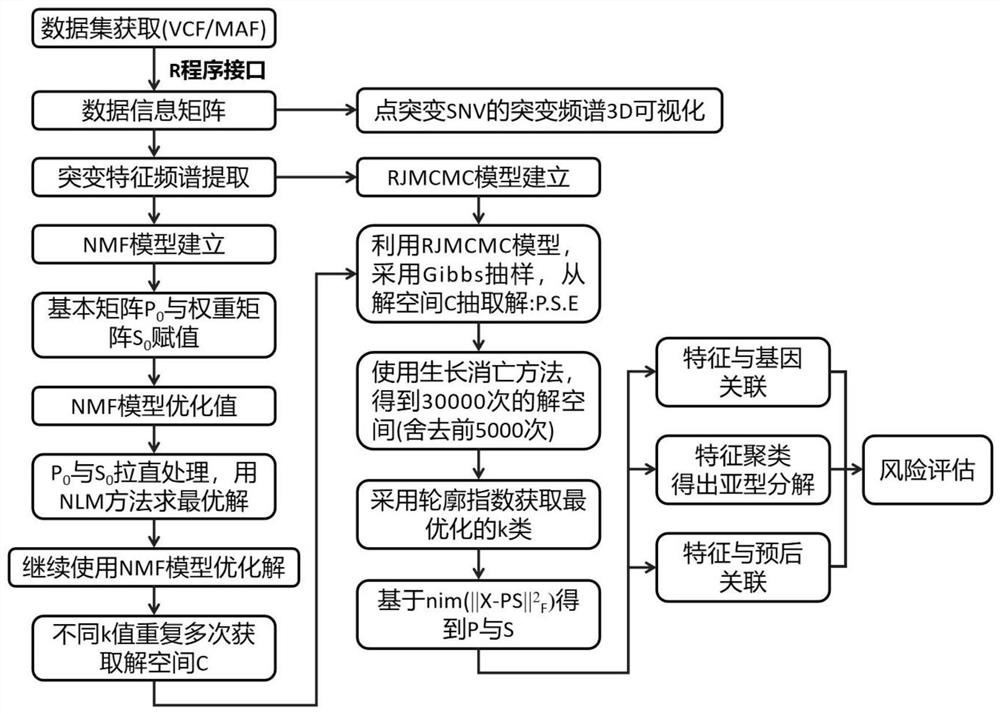
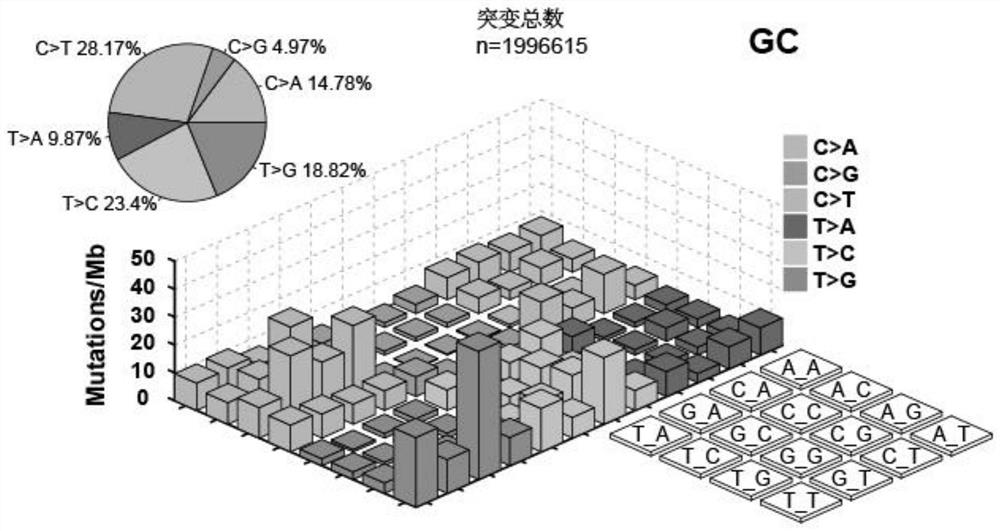
Tumor gene point mutation feature extraction method combining contour coefficient and RJMCMC algorithm
ActiveCN113035275AEnhanced comparison effect displayEasy to useVisual data miningStructured data browsingData setFeature extraction
The invention provides a tumor gene point mutation feature extraction method combining a contour coefficient and an RJMCMC algorithm, and relates to the technical field of tumor gene feature extraction. The tumor gene point mutation feature extraction method combining the contour coefficient and the RJMCMC algorithm comprises the following steps: S1, data set acquisition: mutation types of a mutation data set comprise Somic SNV and Somic INDEL, and the Somic SNV and the Somic INDEL are subjected to overall statistics by using MuTect software; According to the tumor gene point mutation feature extraction method combining the contour coefficient and the RJMCMC algorithm, an input mode of annotation files is realized, use is convenient, early-stage data processing time is saved, the efficiency is improved, mutation spectrum 3D visual display is realized, researchers can visually see mutation conditions of each type from spatial vision, the comparison effect display of the type is is enhanced, and the contour coefficient is innovatively combined to construct the RJMCMCNMF model and algorithm implementation;code software device design is completed, and a software device for feature spectrum and gene association acquisition is achieved.
Owner:GUANGDONG POLYTECHNIC NORMAL UNIV
Method for acquiring anti-false smut gene associated resistance gene analogs and molecular markers thereof
InactiveCN103333884AMark accuratelyIncrease resistanceDNA preparationDNA/RNA fragmentationBiotechnologyGermplasm
The invention discloses a method for acquiring anti-false smut gene associated resistance gene analogs and molecular markers thereof. The method comprises steps of: designing RGA primers for complete genome resistance gene analogs to obtain RGA genotypes of rice germplasms, acquiring 137 anti-false smut phenotypes through a false-smut pathogen microinjection method, combining the RGA genotypes and the anti-false smut phenotypes to perform molecular character association analysis and to perform clone sequencing on PCR products, thus obtaining the resistance gene analogs associated with the anti-false smut gene and the molecular markers thereof. According to the invention, by means of genetic transformation and hybridization, the anti-false smut gene associated resistance gene analogs can be introduced into common cultivated rice varieties to improve resistance of the rice varieties to false smut. The anti-false smut gene associated molecular markers can be used for detecting the existence of the anti-false smut genes, indirectly selecting disease-resistant plants, and protecting identification of inoculated false smut pathogens from being influenced by climate, geographical positions and other uncertain factors, and therefore the anti-false smut gene associated molecular markers can be used for selecting early generation, shortening anti-false smut variety breeding periods, and improving breeding efficiency.
Owner:CROP INST SICHUAN PROVINCE ACAD OF AGRI SCI
Co-detection and association of multiple genes from the same genome in a sample
The present invention is concerned with PCR-based detection methods and kits for the identification, differentiation, and quantification of different bacterial strains (e.g., Gram-negative bacterial strains), and also association of two or more PCR-positive genes to a single genome. The methods generally comprise carrying out PCR reactions using at least a first PCR primer set and / or probe for at least one target nucleic acid; and a second PCR primer set and / or probe for at least a second target nucleic acid. Positive PCR reaction products are then detected to determine test samples containing positive PCR reaction products for both the first and second target nucleic acids. This information can be used to calculate the gene association rate to determine whether the sample contains, for example, Shiga toxin-producing E. coli of the O-type serogroup.
Owner:KANSAS STATE UNIV RES FOUND
Integrating method of multiple kinds of biological sequence notes
ActiveCN110223732AIncrease profitExcellent quantitative abilitySequence analysisInstrumentsTranscription startMedical treatment
The invention discloses an integrating method of multiple kinds of biological sequence notes. The method comprises the steps of extracting one kind of biological sequencing data from biological sequencing data as a main biological sequence set, and using the rest biological sequencing data as an auxiliary biological sequence set; establishing a sequence-gene association mapping set; acquiring a basic association area and an extended association area of the gene according to a gene transcription starting point; for the sequence of the main biological sequence set, performing traversal on the extended association area of the gene, and if the area with the sequence is intersected with the extended association area of a certain gene, establishing a sequence-gene association mapping between thegene and the sequence; applying reference data on a biological sequence noting result in the sequence-gene association mapping set and calculating the significance by means of ultra-geometric testingand binomial testing; respectively sequencing the notes which are obtained by the two methods, adding the sequence numbers of the same notes, performing sequencing again and using the sequencing result as a note result of multiple kinds of biological sequence data. The integrating method realizes noting of various characteristics and has application value in the medical field.
Owner:TSINGHUA UNIV
Method and system for network modeling to enlarge the search space of candidate genes for diseases
ActiveUS10347359B2Biological testingSystems biologyGenomic sequencingCandidate Gene Association Study
With the advent of low cost, high-throughput whole genome sequencing (“next generation sequencing”), tools are available to assay human genetic variation contributing to inherited disease syndromes. A method is disclosed for prioritization of genetic variants, and identification of disease genes, using network modeling of gene associations.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
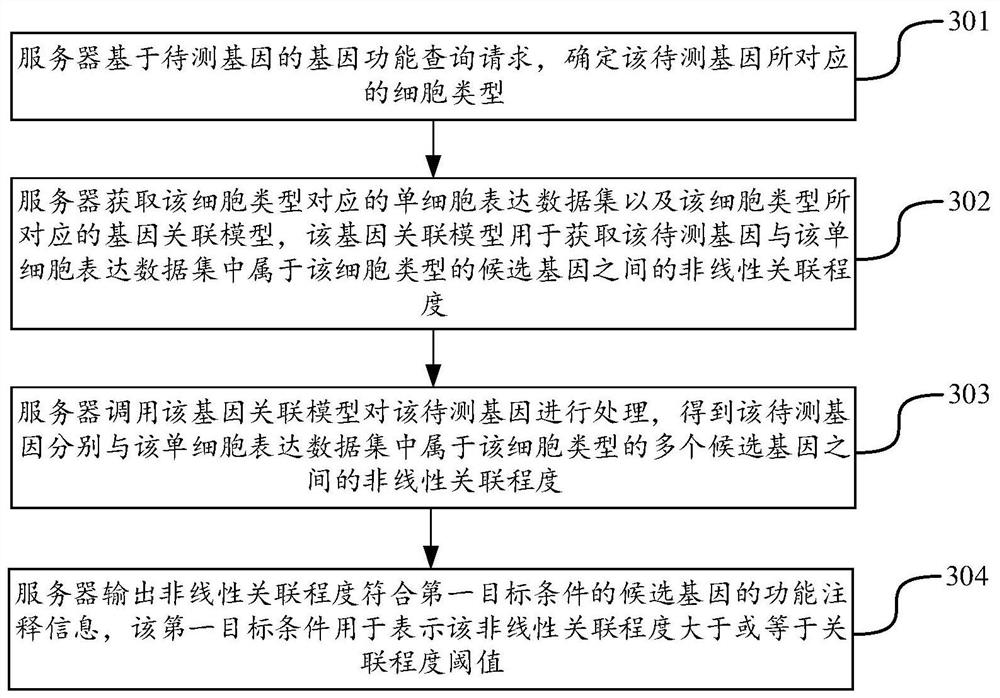
Gene data processing method and device, computer equipment and storage medium
PendingCN114765062AImprove accuracyProteomicsGenomicsCandidate Gene Association StudyLinear relationship
The invention discloses a gene data processing method and device, computer equipment and a storage medium, and belongs to the technical field of computers. According to the method, the gene association model corresponding to the cell type to which the to-be-detected gene belongs can be called through the gene function query request of the to-be-detected gene, and the nonlinear association degree between the to-be-detected gene and the known candidate gene is mined; the candidate genes with high nonlinear correlation degree are used for labeling the function annotation information of the to-be-detected genes, the mode of extracting the nonlinear relation by calling the gene correlation model is completely different from the mode of extracting the linear relation in the traditional statistics, the candidate genes with higher similarity with the to-be-detected genes can be deeply mined, and the accuracy of the to-be-detected genes is improved. The similarity is implicit non-linear similarity instead of non-linear similarity, so that the accuracy of the gene data processing process is greatly improved.
Owner:TENCENT TECH (SHENZHEN) CO LTD
A method for gene association analysis based on deep learning algorithm
Owner:HANGZHOU DIANZI UNIV
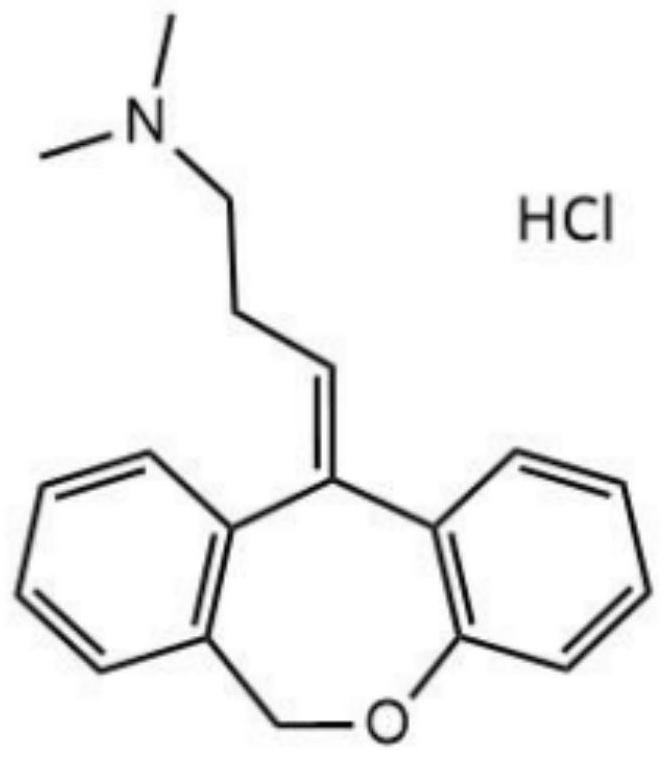
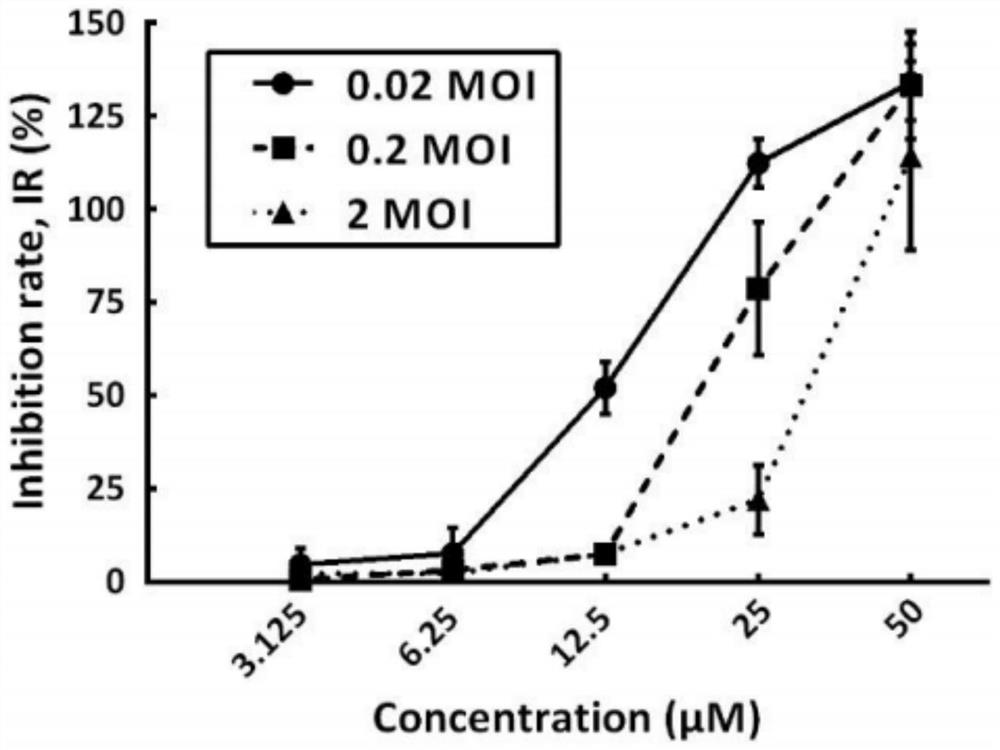
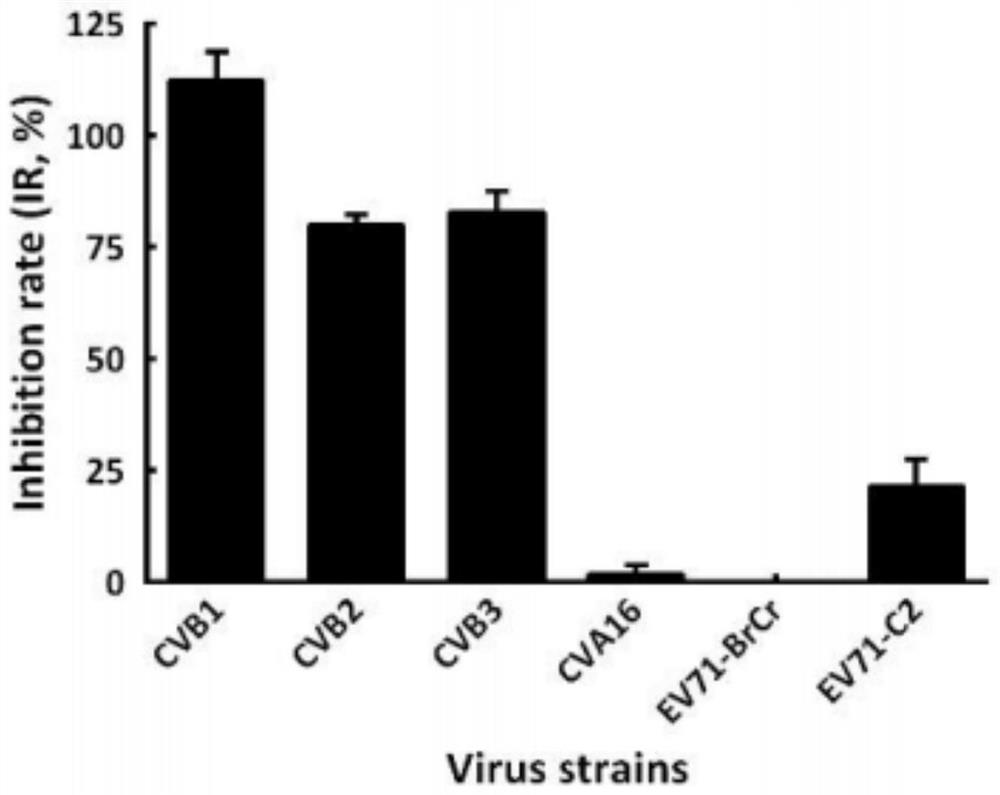
Application of doxepin hydrochloride in preparation of antiviral drugs
ActiveCN114344288AStrong antiviral activityInhibition of replicationOrganic active ingredientsAntiviralsCoxsackie VirusesAntiviral drug
The invention provides an application of doxepin hydrochloride in preparation of antiviral drugs, and relates to the technical field of medicines, research shows that doxepin hydrochloride shows relatively strong antiviral activity to coxsackie B viruses type 1, type 2 and type 3, the minimum 50% inhibitory concentration (IC50) value is 10.12 + / -0.85 mu M. In addition, it is found that doxepin hydrochloride inhibits virus replication in the early stage of an infection cycle, and doxepin hydrochloride can inhibit virus replication in the early stage of the infection cycle. Meanwhile, some host genes related to a mechanism are found through target spot prediction and gene association network analysis, and foundation and basis are provided for doxepin hydrochloride used for treating coxsackie type B virus infection.
Owner:SHENZHEN TECH UNIV
An Integrative Method for Multi-Class Biological Sequence Annotation
ActiveCN110223732BIncrease profitExcellent quantitative abilitySequence analysisInstrumentsSequence annotationComputer science
The invention discloses a method for integrating annotations of multiple types of biological sequences, comprising: selecting one biological sequencing data from biological sequencing data as a main biological sequence set, and the rest as auxiliary biological sequence sets; establishing a sequence-gene association mapping set; According to the gene transcription start point, the basic association region and the extended association region of the gene are obtained; for the sequence of the main biological sequence set, the extended association region of the gene is traversed, and if the region where the sequence is located overlaps with the extended association region of a gene, then the Sequence-gene association mapping of genes and sequences; the hypergeometric test and binomial test are used to calculate the significance of the results of reference data applied to the biological sequence annotations in the sequence-gene association mapping set; the annotations obtained by the two methods are sorted separately, and Add up the sequence numbers of the same annotations and then sort them again as the annotation results of various biological sequence data. The invention realizes the annotation of various features, and has application value in the medical field.
Owner:TSINGHUA UNIV
Identity elucidation of unknown metabolites
A method of elucidating the identity of an unknown metabolite comprising measuring amounts of known and unknown metabolites in subjects; associating the unknown metabolite with a specific gene from a gene association study; determining a protein associated with the specific gene and analyzing information for the protein; associating the unknown metabolite with concentrations and / or ratios of other metabolites in subjects; obtaining chemical structural data for the unknown metabolite; and using the information obtained to elucidate the identity of the unknown metabolite.
Owner:METABOLON +1
Genotype-phenotype association analysis method in multi-omics data based on small sample
ActiveCN113192556BSolve the problem that the eigenvalues are too large to be effectively regressedImprove forecast accuracyBiostatisticsProteomicsGenotypeGene association
Owner:NORTHWESTERN POLYTECHNICAL UNIV +1
Use method of haplotype ancestral database
The invention discloses a use method of a haplotype ancestral database. The use method comprises the following steps of: moving an identification frame to identify information of n continuous SNP (Single Nucleotide Polymorphism) sites falling into the identification frame; matching the information of n continuous SNP loci in the recognition frame with the comparison units, and if the comparison units with the same information of the SNP loci cannot be matched, determining that the SNP loci are unknown; if a comparison unit with the same information of the SNP sites can be matched, recording corresponding race information of the comparison unit, calling an identification frame capable of identifying n + 1 continuous SNP sites, and identifying the n + 1 continuous SNP sites containing the n SNP sites which are the same as the comparison unit; and repeating the previous step until the information of a plurality of continuous SNP sites in the identification frame cannot be matched with the comparison unit with the same SNP site information. The method has the advantages that the ancestor tracing speed is high, the error is small, the definition is high, the big data capacity is large, and a basis is provided for finding out disease and gene association, precise medical treatment, molecular detection, medication guidance and other technical researches in the medical field.
Owner:广州鸿溪见杉科技有限公司
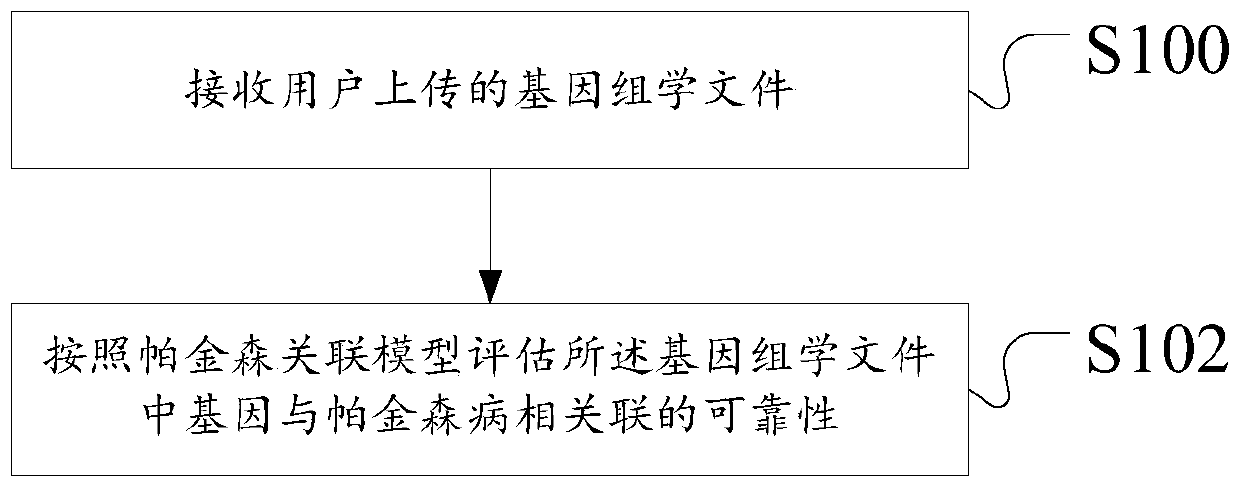
Management method and device based on Parkinson's disease genomics association model
The invention discloses a management method based on a Parkinson's disease genomics association model. The method comprises the following steps: receiving a genomics file uploaded by a user on a terminal; and evaluating the reliability of the gene associated with Parkinson's disease in the genomics file according to a Parkinson's disease association model. According to the method and the device, the technical problems of complex and slow operation and weak user selectivity caused by incapability of realizing intelligent judgment of Parkinson's disease and gene association are solved.
Owner:XIANGYA HOSPITAL CENT SOUTH UNIV
Co-detection and association of multiple genes from the same genome in a sample
The present invention is concerned with PCR-based detection methods and kits for the identification, differentiation, and quantification of different bacterial strains (e.g., Gram-negative bacterial strains), and also association of two or more PCR-positive genes to a single genome. The methods generally comprise carrying out PCR reactions using at least a first PCR primer set and / or probe for at least one target nucleic acid; and a second PCR primer set and / or probe for at least a second target nucleic acid. Positive PCR reaction products are then detected to determine test samples containing positive PCR reaction products for both the first and second target nucleic acids. This information can be used to calculate the gene association rate to determine whether the sample contains, for example, Shiga toxin-producing E. coli of the O-type serogroup.
Owner:KANSAS STATE UNIV RES FOUND
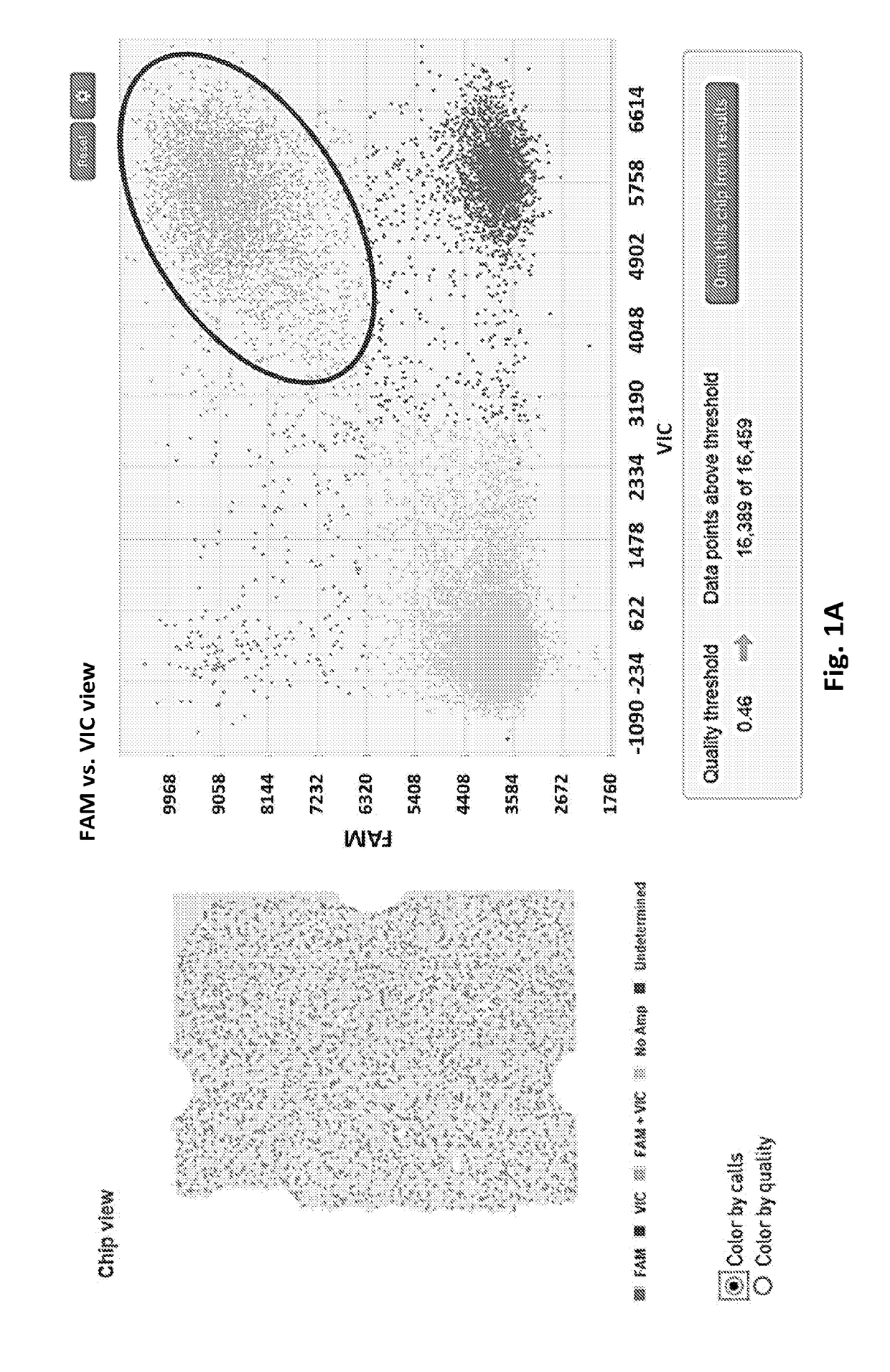
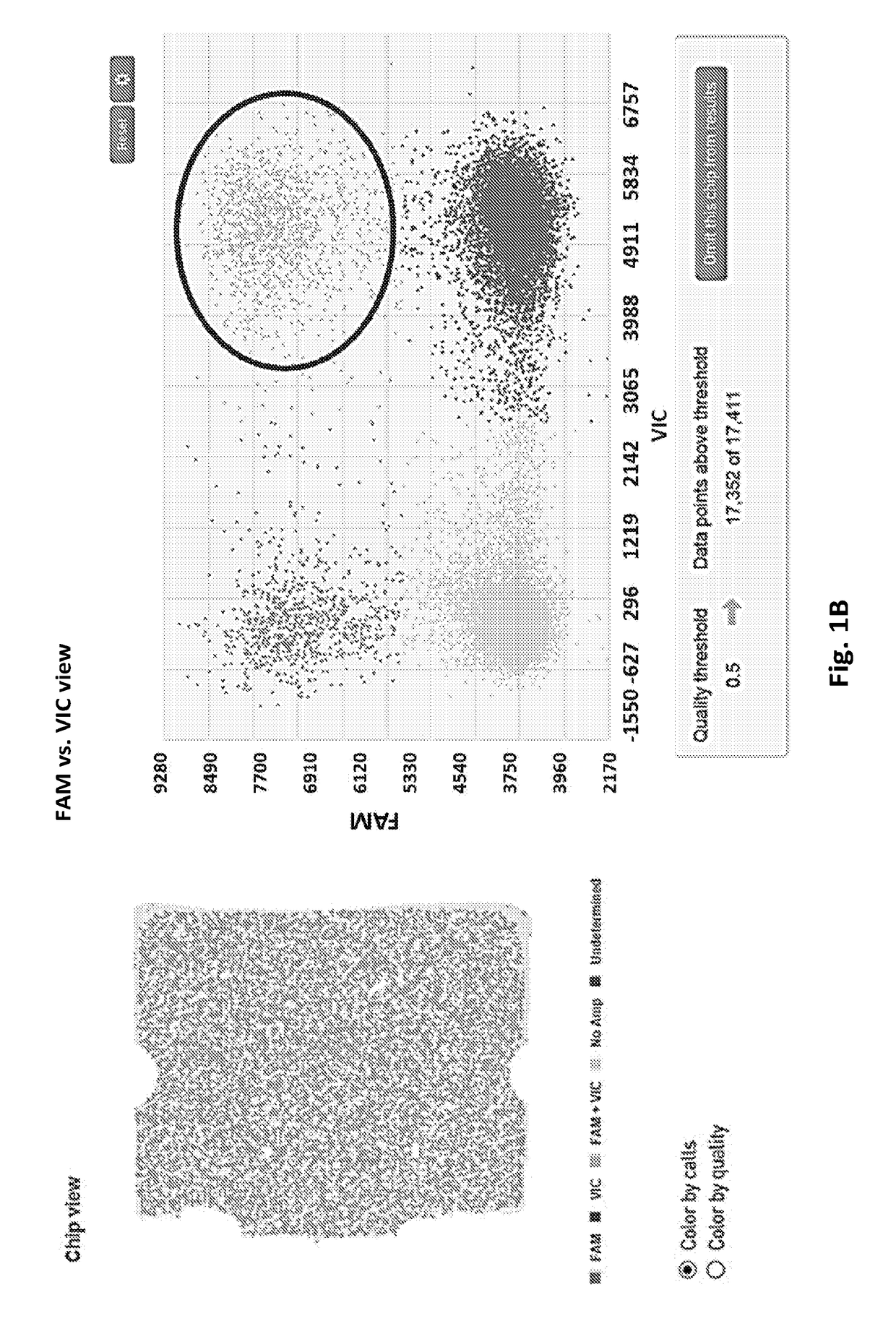
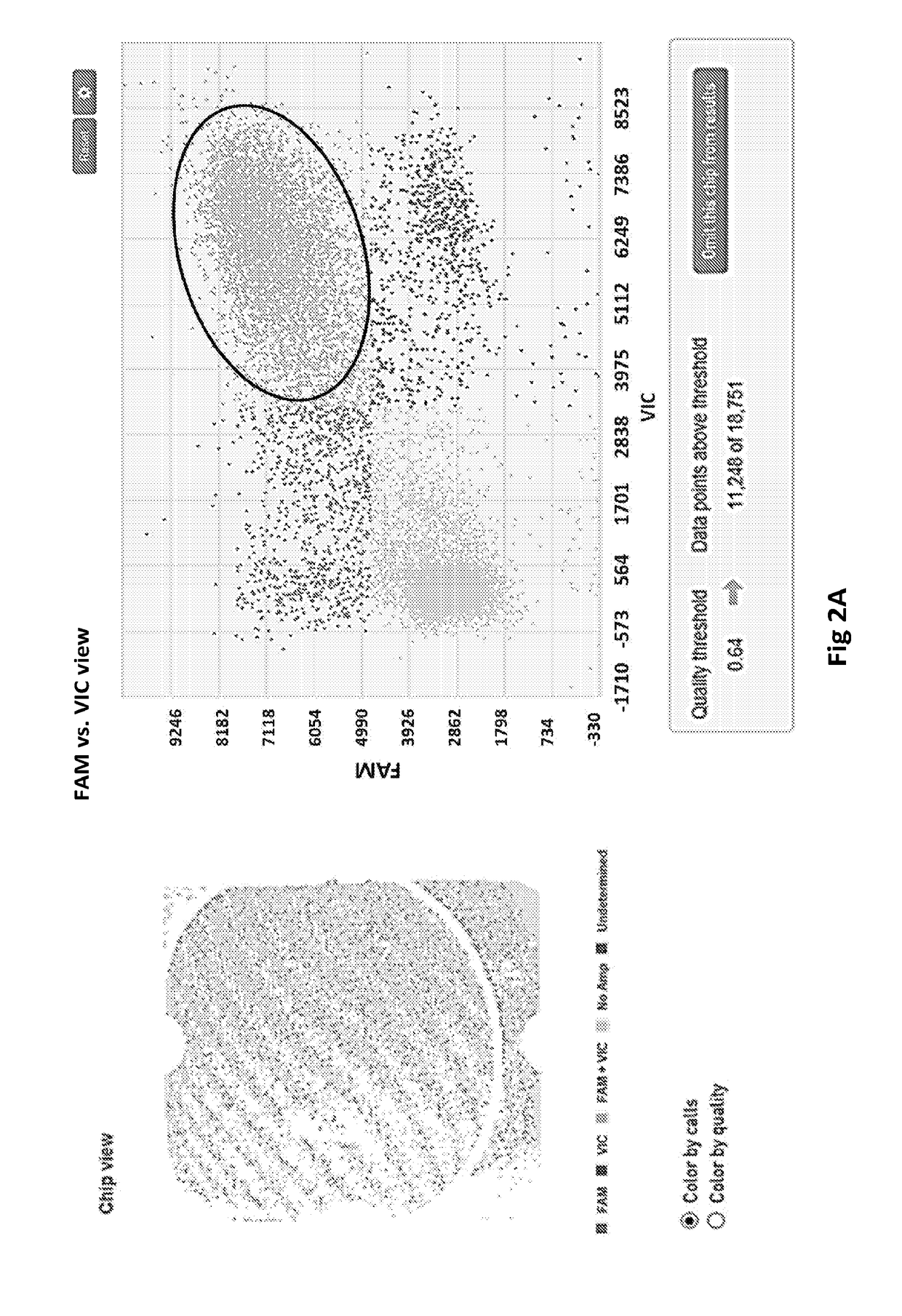
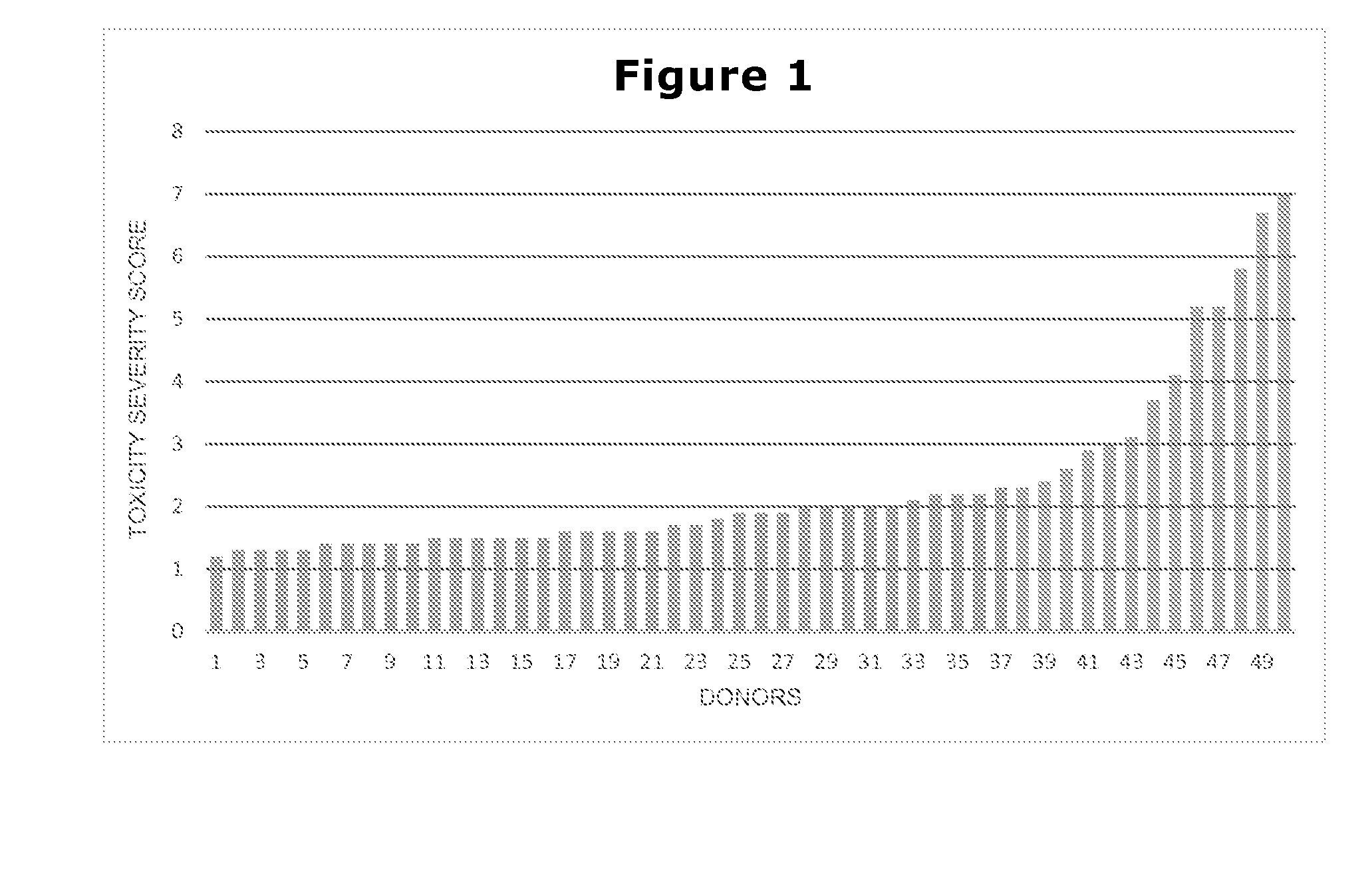
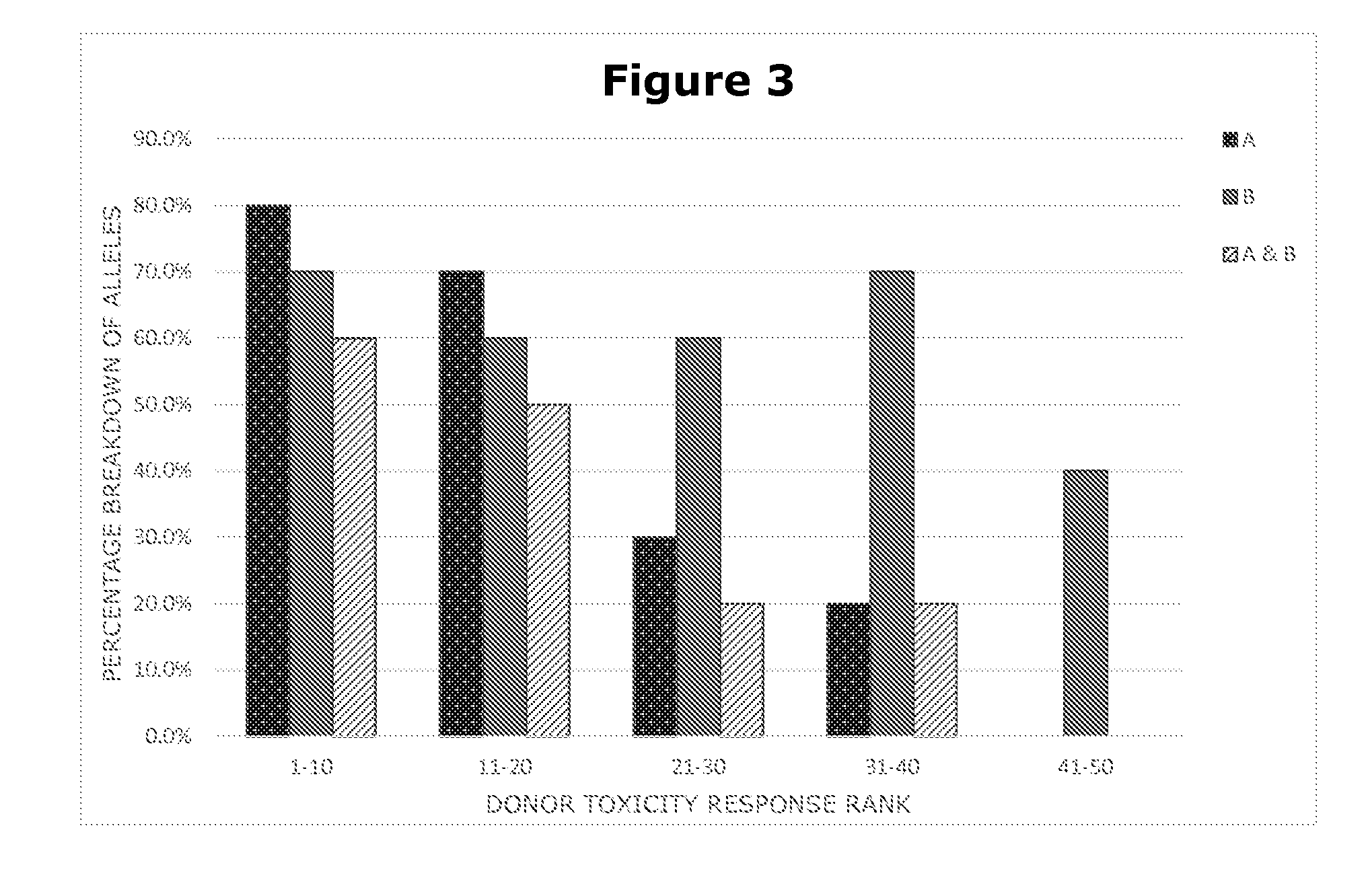
Methods for Genetically Diversified Stimulus-Response Based Gene Association Studies
InactiveUS20160195514A1Improve abilitiesLibrary screeningLibrary member identificationGenetic diversityStudy methods
Methods are provided for improving the impact of genetically diversified stimulus response gene association (GDSRGA) studies. The methods may involve developing subpopulations to be contrasted in GDSRGA studies by obtaining a biological sample from each donor of a population of donors; selecting a common cohort from the biological samples by obtaining at least a partial genomic sequence from each biological sample, aligning the sequences of the biological samples, and removing biological samples that cannot be sequenced accurately or fail to align; applying a test molecule or condition to the biological samples to induce phenotypically distinct responses among the members of the cohort; and segregating the biological samples into subpopulations based on the phenotypically distinct responses. These subpopulations may be used in GDSRGA studies.
Owner:COYNE SCI
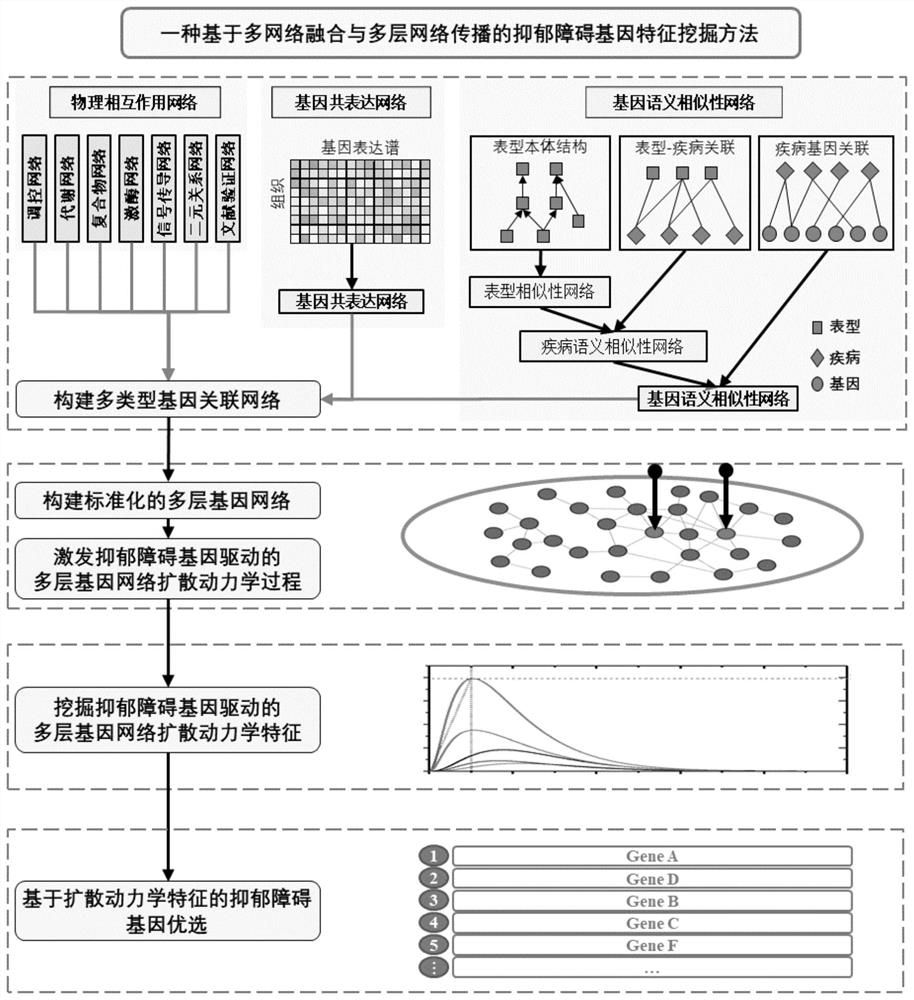
A genetic feature mining method for depressive disorders based on multi-network fusion and multi-layer network diffusion
ActiveCN112837752BStrong discriminationEfficient miningMedical data miningBiostatisticsGene driveData mining
The invention discloses a depressive disorder gene feature mining method based on multi-network fusion and multi-layer network diffusion. The mining method mainly includes the following steps: 1. Construct a multi-type gene association network; 2. Construct a standardized multi-layer gene network; 3. Stimulate the diffusion dynamics process of the multi-layer gene network driven by the depressive disorder gene; 4. Mining the diffusion dynamics of the multi-layer gene network driven by the depressive disorder gene. This mining method can effectively integrate different types of biomolecular networks, and mine effective disease gene characteristics from various diverse biomolecular networks, so as to identify depression-related genes more effectively.
Owner:CENT SOUTH UNIV
Small sample-based genotype and phenotype association analysis method in multi-omics data
ActiveCN113192556ASolve the problem that the eigenvalues are too large to be effectively regressedImprove forecast accuracyBiostatisticsProteomicsGene associationProtein
The invention discloses a small sample-based genotype and phenotype correlation analysis method in multi-omics data. The method specifically comprises the following steps: generating a weighted undirected gene association graph by using a protein network and a gene expression value, and clustering the undirected graph by using an SPICi clustering method to generate a gene cluster; screening the gene clusters by using a group Lasso method; obtaining an SNP cluster corresponding to the screened gene cluster through the eQTL data; constructing each SNP cluster, the corresponding gene cluster and the phenotype into a three-layer network class block, performing regression operation on the association relationship between the SNP and the gene in each class block by adopting a sparse partial least square method, and performing operation on the association relationship between the gene and the phenotype by adopting logistic regression; and averaging the obtained prediction results of the blocks to obtain a final prediction result. The method can solve the problem that effective regression cannot be realized due to huge characteristic values under the condition of small samples in a three-layer network; wherein the prediction accuracy is improved, the biological significance is clearer and tissue specificity is considered.
Owner:NORTHWESTERN POLYTECHNICAL UNIV +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com