Method for identifying litchi varieties by using wide targeted metabonomics technology
A metabolomics and variety technology, applied in the field of litchi variety identification, can solve the problems of low efficiency, hindering substantial breakthroughs in molecular marker-assisted selection breeding technology, and poor reliability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
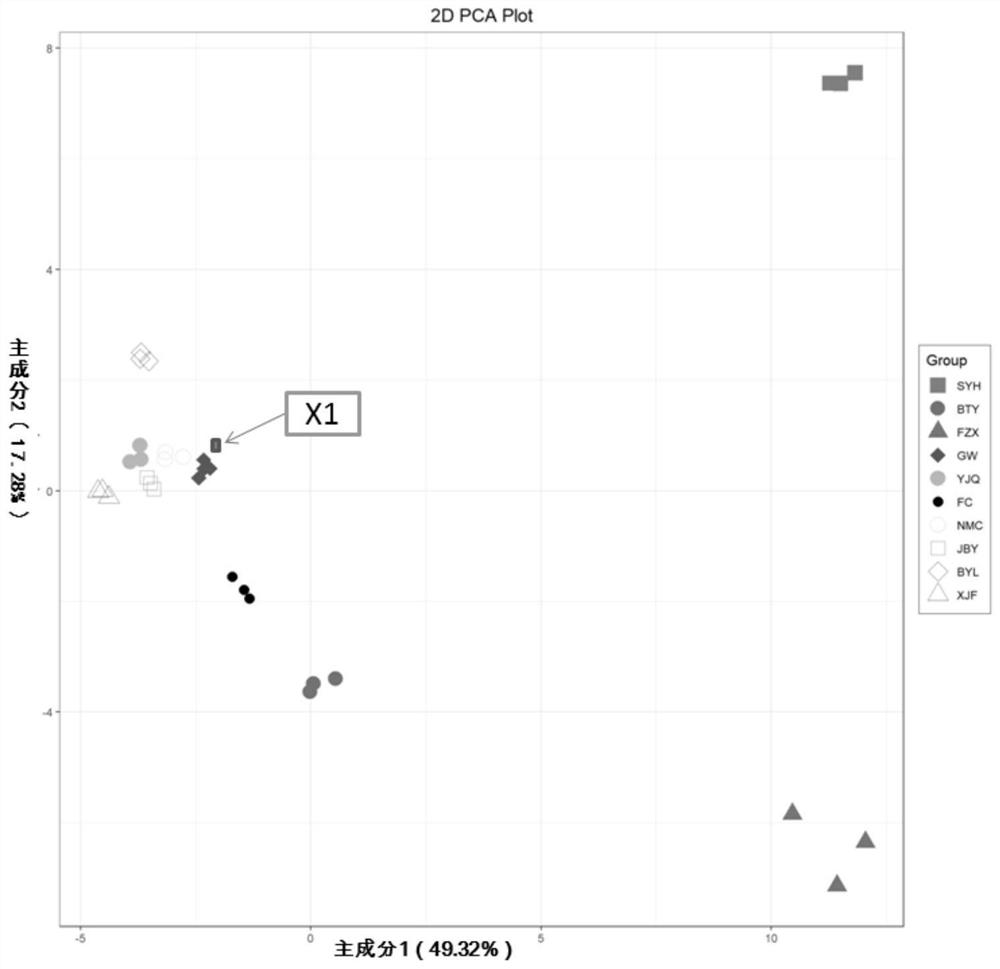
[0042] Taking the Identification of "Guiwei" Litchi as an Example
[0043] 1. Mass spectrometric identification of metabolites in litchi variety library, construction of peak area database, screening of differential metabolites and construction of heatmap.
[0044] (1) Preparation of lychee samples, the process is as follows: 10 known conventional commercial varieties ("Baitangying", "Feizixiao", "Feicui", "Guiwei", "Jinbaoyin", "Nuomici" ", "Sanyuehong", "Xianjinfeng", "Yujinqiu", respectively referred to as BTY, FZX, FC, GW, JBY, NMC, SYH, XJF, YJQ in this application) litchi samples in the mature period ( Maturity is 100%) after picking, the pulp is cleaned, stripped, vacuum freeze-dried, and ground to powder with a grinder; 100 mg of powder is accurately weighed and dissolved in 1.0 mL of 70% (v / v) methanol extract; The dissolved samples were vortexed rapidly 3 times, 5min each time, sonicated once (350W, 35kHz), and refrigerated overnight at 4°C; after refrigerated centr...
Embodiment 2
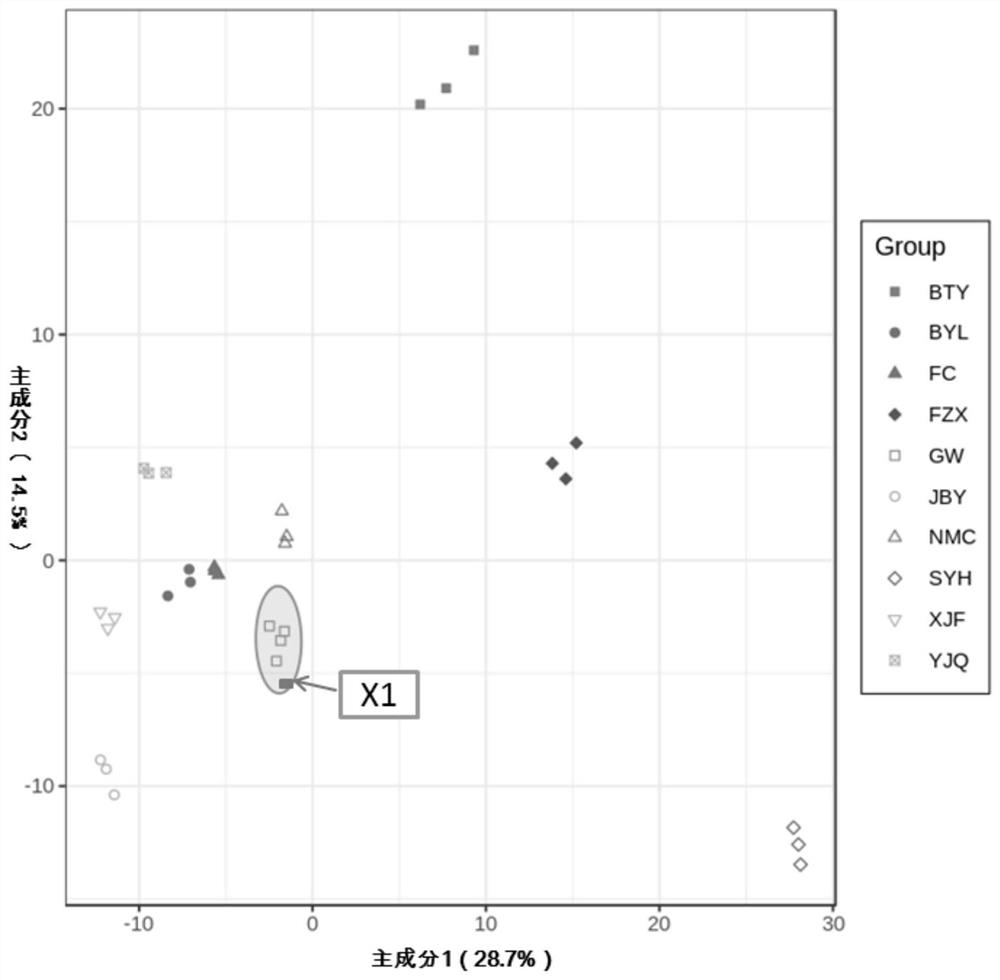
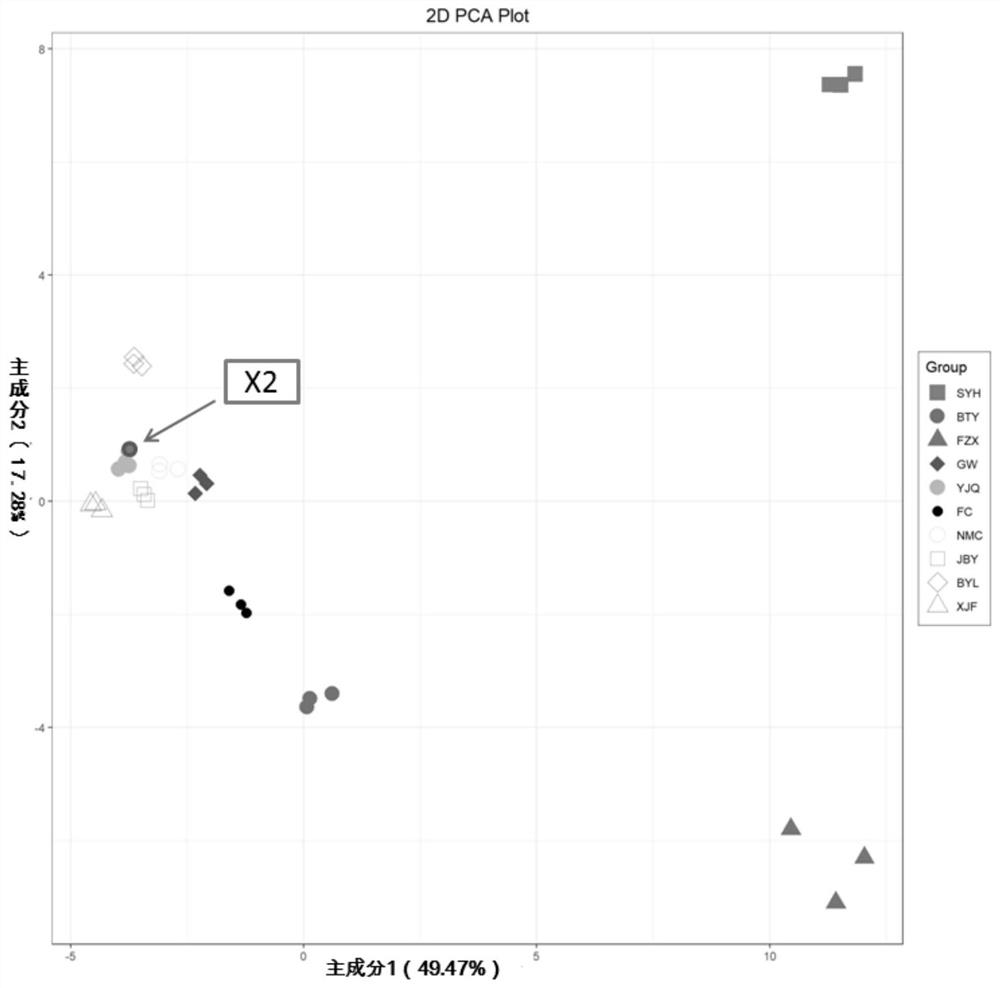
[0055] Take "Yujinqiu" litchi as an example
[0056] 1. Mass spectrometric identification of metabolites in litchi variety library, construction of peak area database, screening of differential metabolites and construction of heatmap.
[0057] (1) The preparation of lychee sample, process is: after the lychee sample of known variety name (same as embodiment 1) is plucked at the full maturity stage (maturity is 90%), the pulp is cleaned and peeled off to get the pulp, and then vacuum freeze-dried, Use a grinder to grind to powder; accurately weigh 100mg of powder, and dissolve it in 1.0mL70% (v / v,) methanol extract; the dissolved sample is vortexed rapidly 3 times, 5min / time, ultrasonicated 1 time (350W , 35kHz), 4°C refrigerator overnight; after refrigerated centrifugation (rotational speed 10,000g, 10min), absorb the supernatant, filter the sample with a 0.22μm microporous membrane and put it into a sample vial for LC-MS / MS analysis . At least 3 biological replicates were p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com