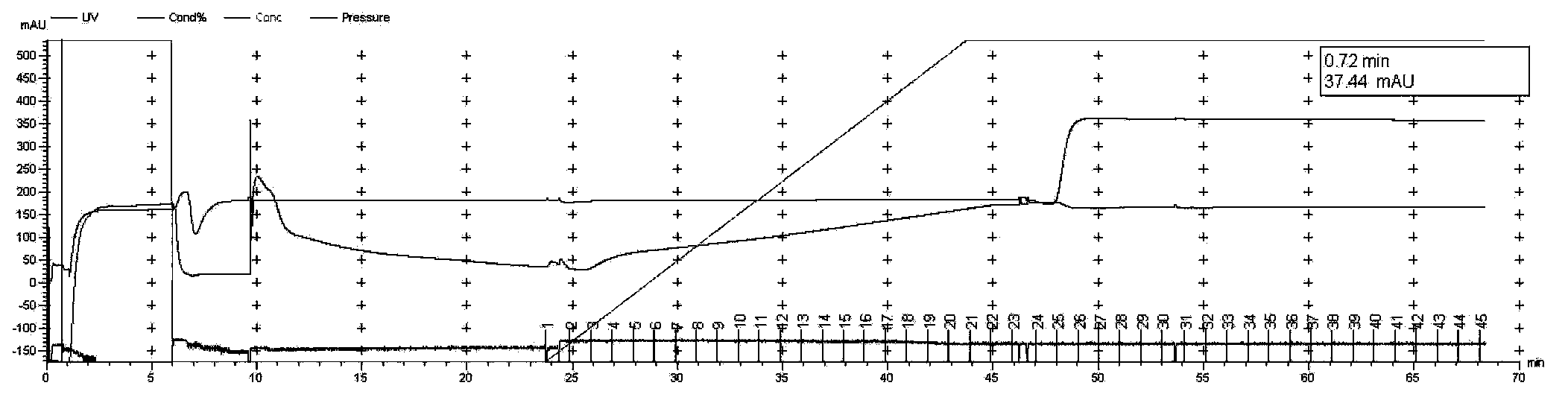Method for identifying ubiquitin-like modification sites of proteins
An identification method and ubiquitin-like technology, applied in the field of proteomics, can solve the problems of inability to provide fragment ions, complexity, and limited collision energy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
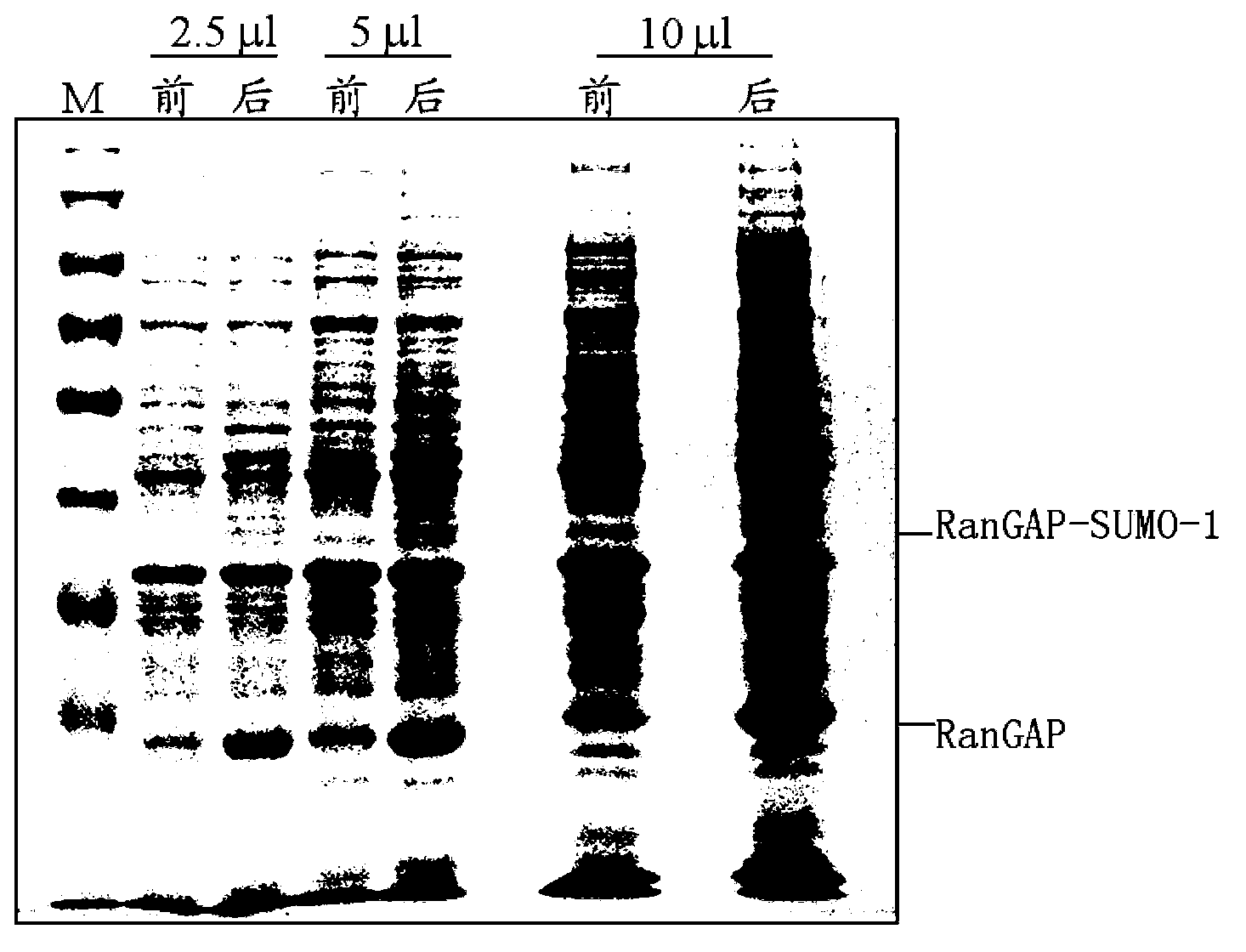
[0081] Example: Identification of Rangap-His-SUMO1-like ubiquitination modification sites
[0082] 1. Target protein purification
[0083] (1) Construct the co-expression plasmid containing the RanGap fragment of Xenopus laevis and human His-SUMO1 (hereinafter referred to as Rangap-His-SUMO1), inoculate 25ml LB medium with 25μl bacteria, and inoculate the bacteria solution to 2× at 1:100 Shake YT medium, 2L / bacteria, add corresponding antibiotics, 280rpm, 37°C, test the OD600 of the bacterial solution after about 2-3 hours, wait until the temperature of the bacterial solution drops to room temperature about 20°C, add IPTG induction, 180rpm, 25°C, 12h.
[0084] The bacterial protein induced by IPTG was collected, and the same amount of bacterial liquid 2OD was also collected to test whether the protein was induced to express. The 12% SDS-PAGE staining is shown in Figure 2-A, and the protein is expressed in the target molecular weight region, and the band is proved to be Rang...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com