Method for quantitatively detecting components of vigna umbellata and vigna angularis in food by double digital PCR
A quantitative detection and red bean technology, applied in biochemical equipment and methods, microbiological measurement/inspection, etc., can solve the technical requirements of the inspection and supervision of food authenticity, and the difficulty of effectively identifying the ingredients of red bean and red bean, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0041] 1. Preparation of samples and extraction of genomic DNA templates: Take 10 g of samples (red bean, red bean, mixture of red bean and red bean, etc.), grind and homogenize with a grinder at 1800 rpm for 3 minutes.
[0042] Weigh 30mg of the sample into a 1.5mL centrifuge tube, and extract the DNA of the sample using the kit method. The kit can be selected: Kurabo QuickGene DNA extraction kit DT-S, Wizard Genomic DNA purification kit (Promega, A1120), PSS nucleic acid automatic extraction instrument, etc. DNA extraction method. Those skilled in the art are familiar with these DNA extraction methods, so as to extract the DNA of corresponding samples respectively.
[0043] 2. Design and synthesize the primers and probe sequences of the red bean species-specific gene sequence and the primer and probe sequences of the red bean universal gene sequence. The nucleotide sequences of the primers and probes are as follows:
[0044]
[0045] 3. Perform double digital PCR reactio...
Embodiment 1
[0094] Example 1: Verification of the Absolute Limit of Quantification of Copy Number
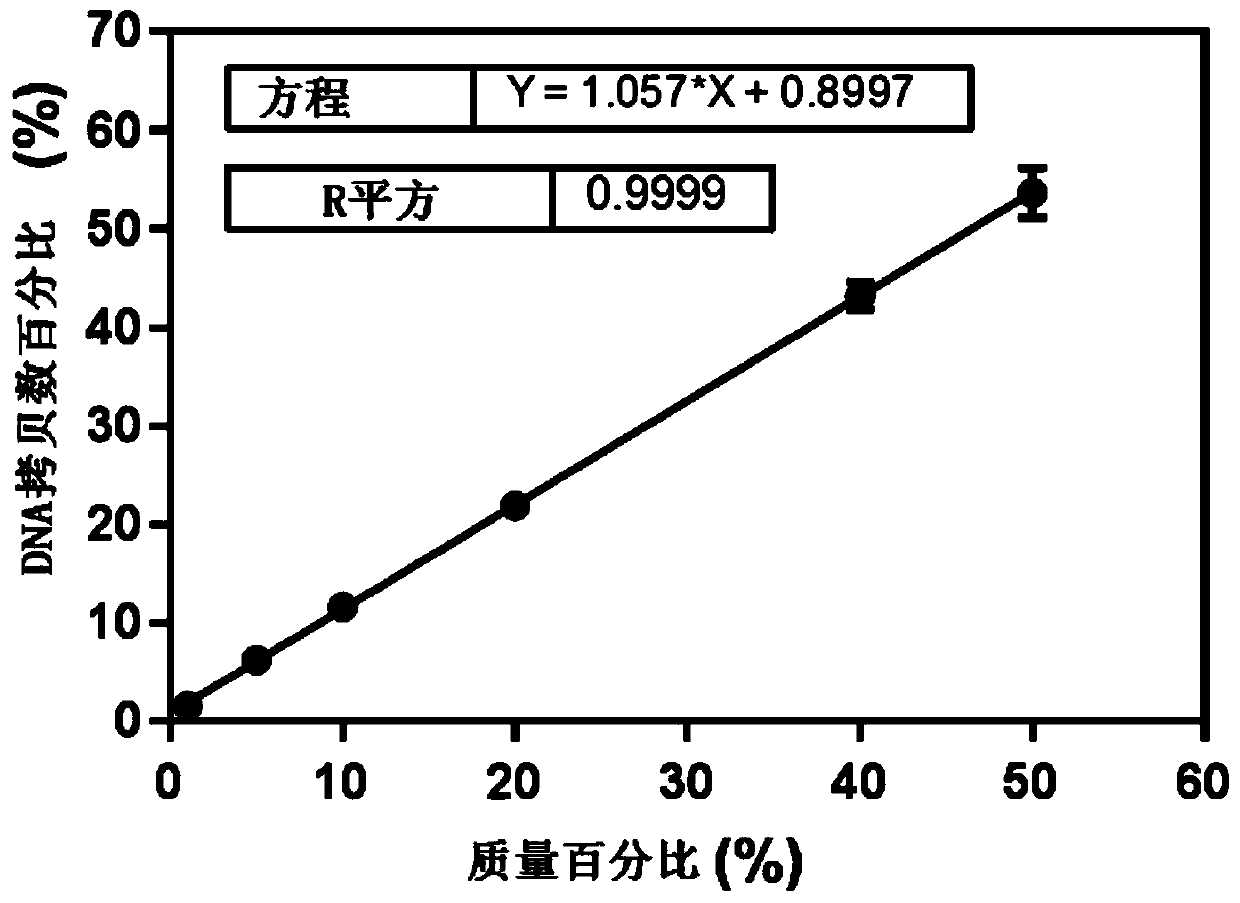
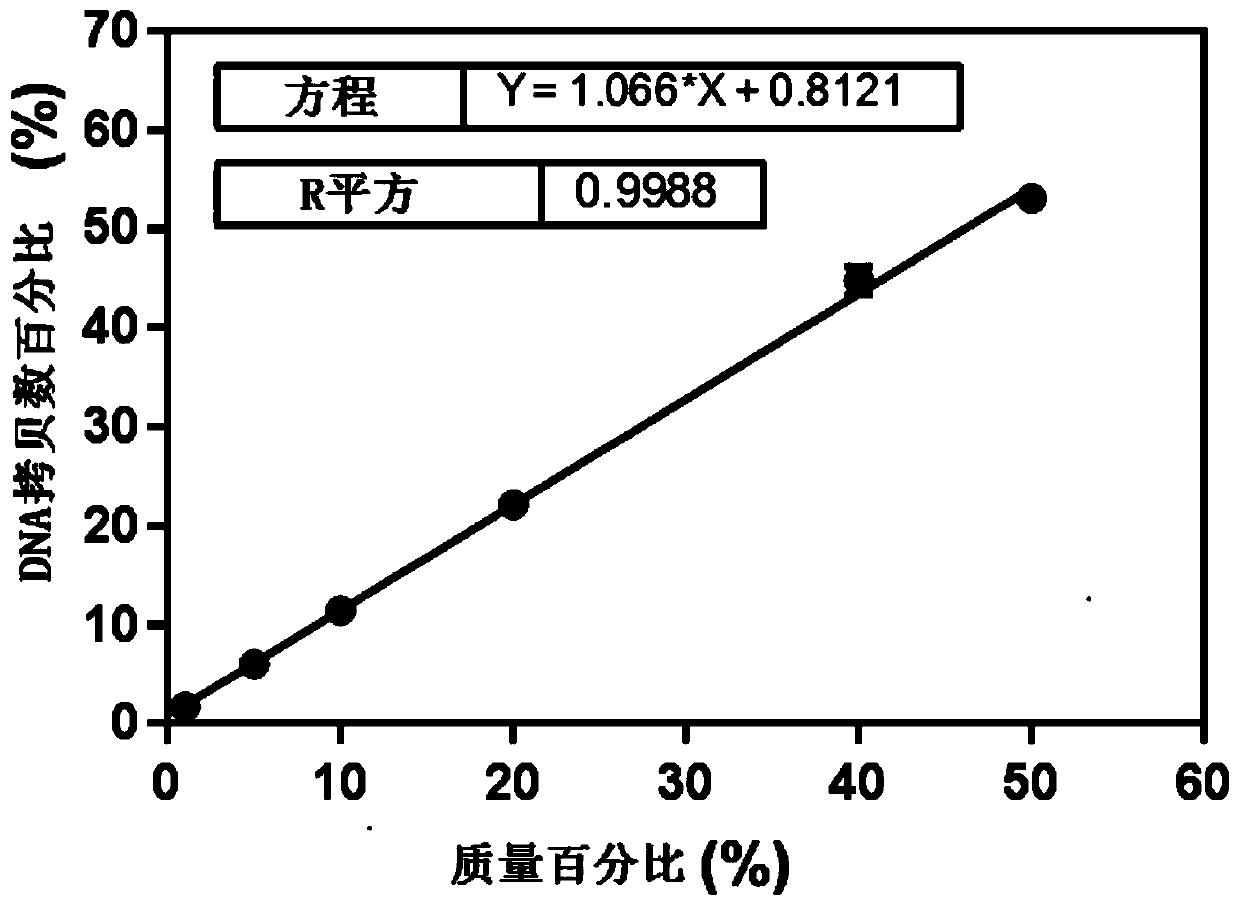
[0095]Samples to be tested: In order to verify the absolute limit of quantification of the copy number of this method, the genomic DNA of red bean and red bean were extracted respectively and serially diluted to obtain red bean DNA at a serial dilution concentration of 300, 100, 20, 10, 5, 1 copy / microliter and Chixiaodou DNA. The red bean species-specific primer probe was used to detect the serially diluted red bean DNA, and the red bean universal primer probe was used to detect the serially diluted red bean and red bean DNA respectively. Three parallel ddPCR and cdPCR experiments were carried out in each experiment. The experimental results are shown in Table 1.
[0096] Table 1 Verification of the absolute quantification limit of DNA copy number in red bean and red bean (ddPCR and cdPCR)
[0097]
[0098] As can be seen from the results shown in Table 1, for red bean-specific genes,...
Embodiment 2
[0101] Example 2: Validation of Quantitative Limit of Copy Number Percentage
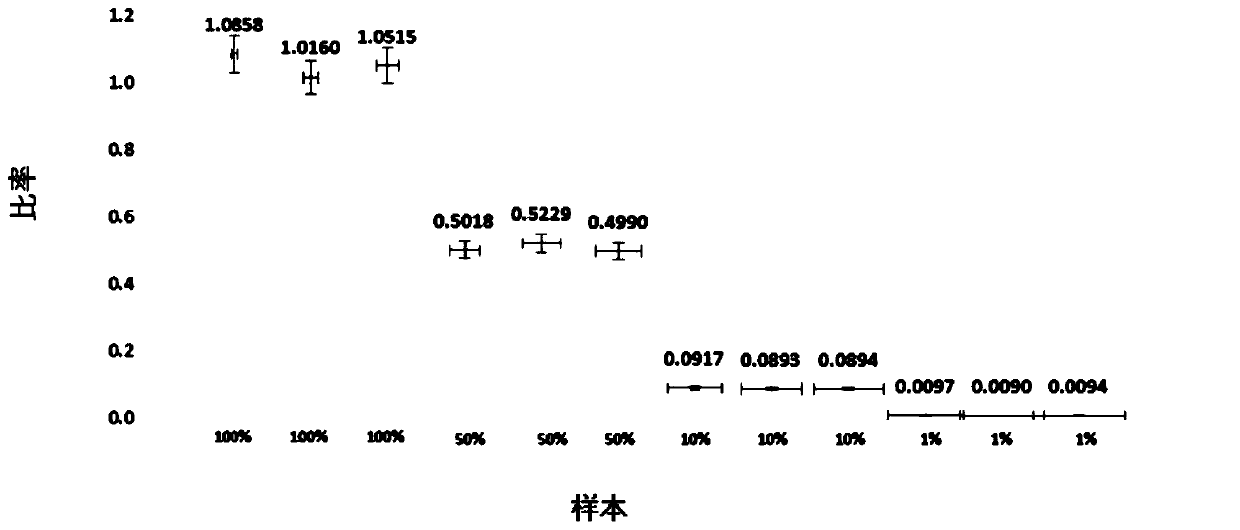
[0102] For the sample: In order to verify the quantitative limit of copy number percentage of this method, the red bean DNA with known copy number concentration was used as the substrate, and the red bean DNA with known copy number concentration was mixed to obtain a series of mixed copy number percentages of 100%, 100%, and 100%, respectively. 50%, 10%, 1%, and 0.1% mixed samples of Adzuki bean and Red bean DNA. Three parallel ddPCR and cdPCR experiments were carried out respectively, and the obtained experimental results are shown in figure 1 and figure 2 .
[0103] Depend on figure 1 and figure 2 From the results shown, it can be seen that for red bean genomic DNA samples whose copy number percentages are 100%, 50%, 10%, and 1%, the detection results on the ddPCR platform are 105.11%, 50.79%, 9.02%, and 0.94%, respectively, The RSD values among the three parallels were between 1.49% and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com