Method for identifying D614G mutation in SARS-CoV-2 based on CRISPR-Cas12a
A D614G, sars-cov-2 technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of complex operation, high difficulty and high cost, and achieve high detection sensitivity, The effect is strong, the method is simple and easy to implement and the effect is fast
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
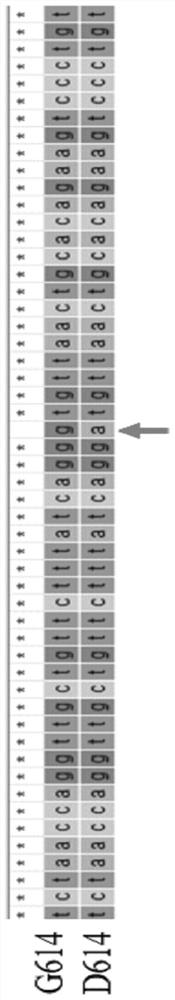
[0033] Example 1. Identification of SARS-CoV-2D614G mutation based on CRISPR-Cas12a
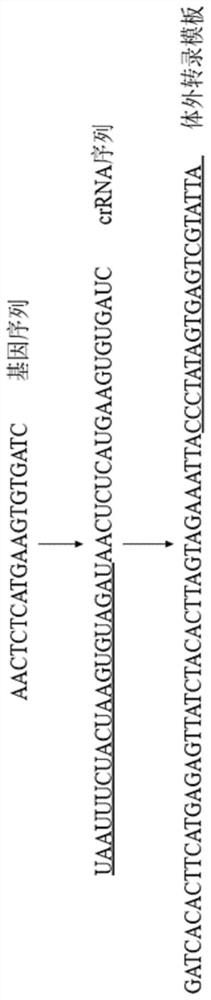
[0034] 1. Find targets containing mutation sites and design target crRNA
[0035] The nucleic acid detection target gene sequence of the CRISPR-Cas12a system needs to have a TTTN / TTN PAM sequence at the 5' end, and select a suitable detection target at the mutation site. After repeated screening and testing, the present invention selects the detection target located at NC_045512.2 (23387bp to 23406bp).
[0036] Design crRNA according to the gene sequence of the target and the CRISPR-Cas12a nucleic acid detection system. The crRNA sequence consists of two parts: the conserved gene sequence at the 5' end (scaffold / repeat part), and the complementary sequence of the target gene sequence at the 3' end. The conserved gene sequence of crRNA is different in different CRISPR-Cas nucleic acid detection systems. Different crRNAs need to be designed for the target. The crRNA sequence can be directly ...
Embodiment 2
[0075] Example 2. SARS-CoV-2D614G mutation identification based on RT-RAA constant temperature amplification and CRISPR-Cas12a
[0076] 1. Design and screening of constant temperature amplification primers for D614G mutation site identification
[0077] (1) Design the constant temperature amplification primers for each detection target
[0078] The design requirements of the RT-RAA constant temperature amplification primers are: the length of the primer is 30-35bp, the 5' end of the primer is an AT base-rich region, and the 3' end of the primer is a CG base-rich region, so as to avoid the formation of hair loss by the primer itself. The clip structure avoids the formation of primer dimers between upstream and downstream primers, and the Tm value of the melting temperature of the primer itself can be ignored. In this experiment, when designing the constant temperature amplification primers, the fragment length of the amplification products should be kept as small as possible w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com