Standard substance for extensive oncogene detection and preparation method and application thereof
A tumor gene and detection standard technology, applied in the field of pan-tumor gene detection standards and their preparation, which can solve the problems of limited number of driver genes and lack of consensus.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
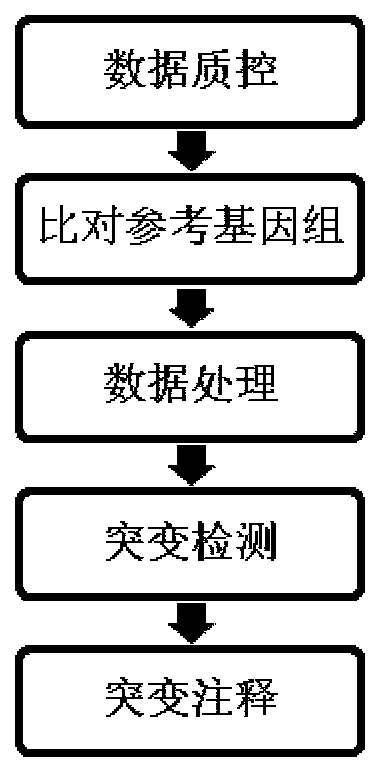
[0078] Example 1 Gene editing and single cell cloning
[0079] The gene editing method described in the present invention can obtain most of the clinically popular tumor gene mutation sites, such as epidermal growth factor receptor gene (EGFR), etc., as shown in the following table:
[0080]
[0081] In one or more embodiments of the present invention, the specific method is:
[0082] a. For the target site of the target gene, design a specific guide RNA (guide RNA, gRNA), and construct the expression vector of the guide RNA. At the same time, single-stranded DNA is designed and synthesized to be used as a template for gene editing and repair.
[0083] b. Co-transfect the expression vector of the guide RNA and the single-stranded DNA into the target cells.
[0084] c. When the cells are in good condition, perform single cell cloning.
[0085] d. After the formation of cell clones, some clones were taken for sanger sequencing to confirm that the gene editing was successfu...
Embodiment 2
[0092] Example 2 Preparation of Pan-tumor Gene Detection Standard Samples to be Detected
[0093] (1) Genomic DNA extraction
[0094] Using Promega 16Cell LEV Purification Kit, Cat. No. AS1140 and the usage method in its instruction manual are used to extract gDNA.
[0095] (2) Concentration determination
[0096] ① Use a spectrophotometer to measure the DNA concentration of the extracted product.
[0097] ② Concentration determination should be repeated three times in a row, and the following conditions should be met:
[0098] Concentration: average concentration 20.0ng / μL≤x≤60.0ng / μL;
[0099] OD260 / 280: The measured value is 1.8≤x≤2.0, which is qualified;
[0100] OD260 / 230: The measured value is 1.5≤x≤5.0, and it is determined to be qualified.
[0101] (3) mixed
[0102] ①Mix the gDNA extracted from the cell pellet with the same code and batch number, and confirm before mixing:
[0103] The gDNA to be mixed is extracted from cell pellets with the same code and bat...
Embodiment 3
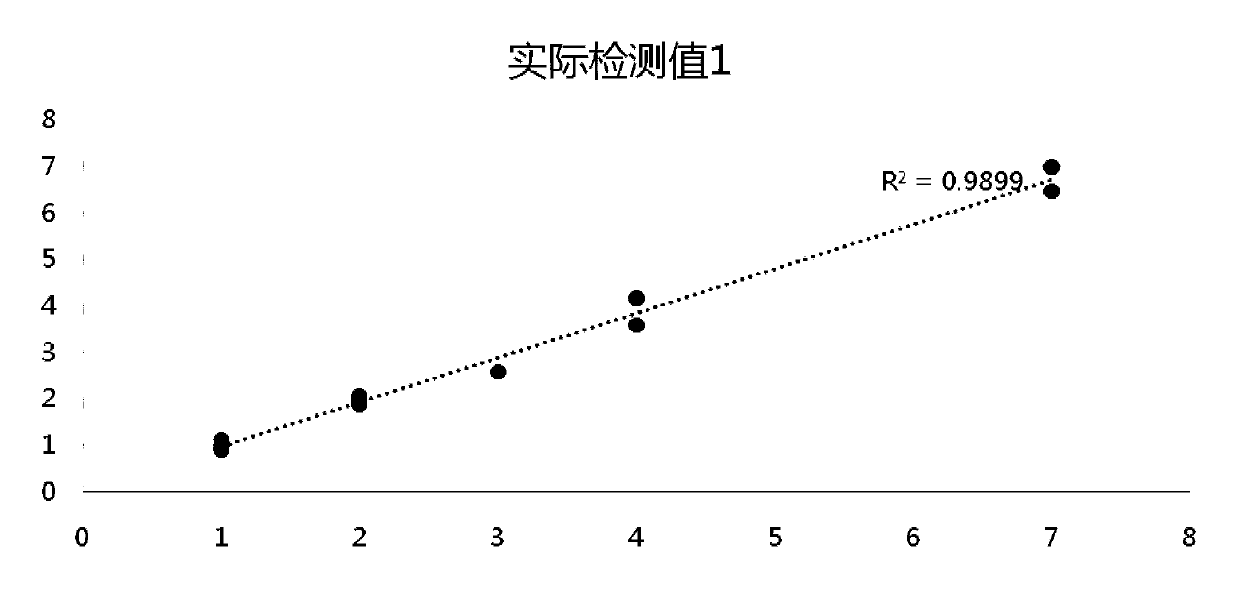
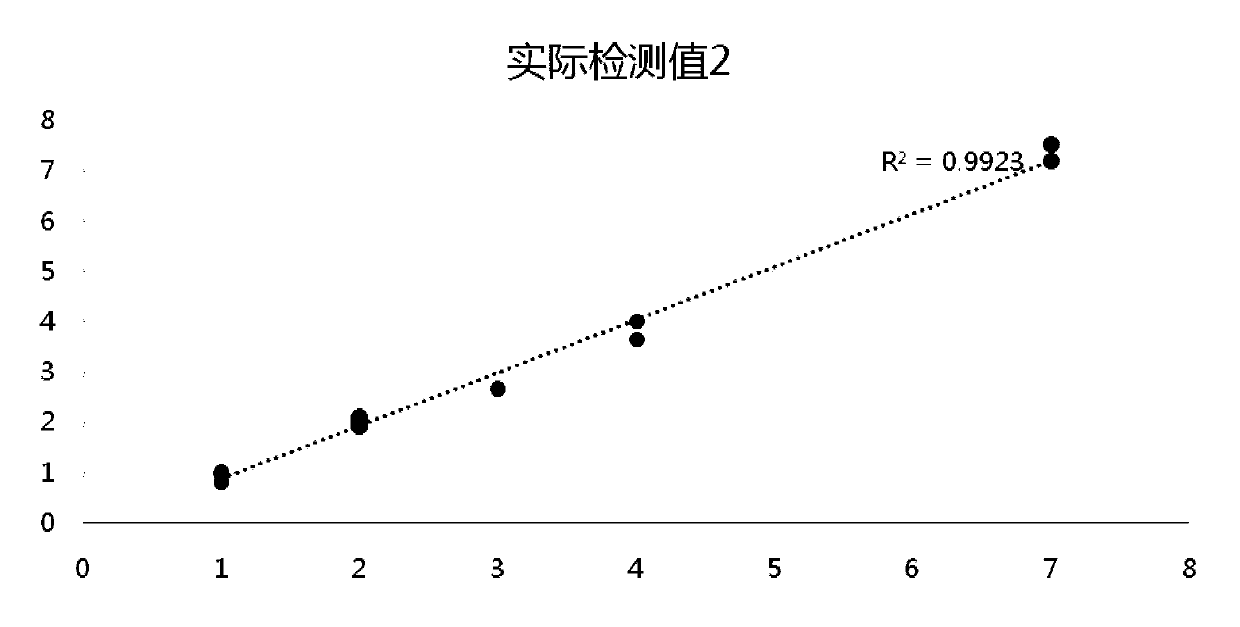
[0112] Example 3 Detection of Pan-tumor Gene Detection Standard Samples to be Detected
[0113] (1) ddPCR detection
[0114] The pan-tumor gene detection standard sample obtained in Example 2 is tested for the gene frequency of the gene loci in the table below by ddPCR, and the original mutation frequency of the raw material is confirmed by ddPCR (BIO-RAD QX200 platform) to obtain ddPCR verification Mutation site variation information.
[0115]
[0116] The specific ddPCR experimental procedure is as follows:
[0117]① Configure the reaction master mix according to the reaction components in the table below, and make 2 to 4 replicate wells for each sample at each site. Considering the loss of pipetting, configure 1.1 reaction mixtures for each amplification well at each site. quantity. See the table below for detailed configuration components and their dosage:
[0118]
[0119] ② Droplets occur. Add the above-mentioned premix to the middle hole of the droplet genera...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com