Method for deleting drug resistant genes of acinetobacter baumannii (AB) through CRISPR-Cas9
A technology of Acinetobacter baumannii and drug-resistant genes, applied in the field of gene editing, to achieve high knockout efficiency, prevent the spread of drug-resistant genes, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
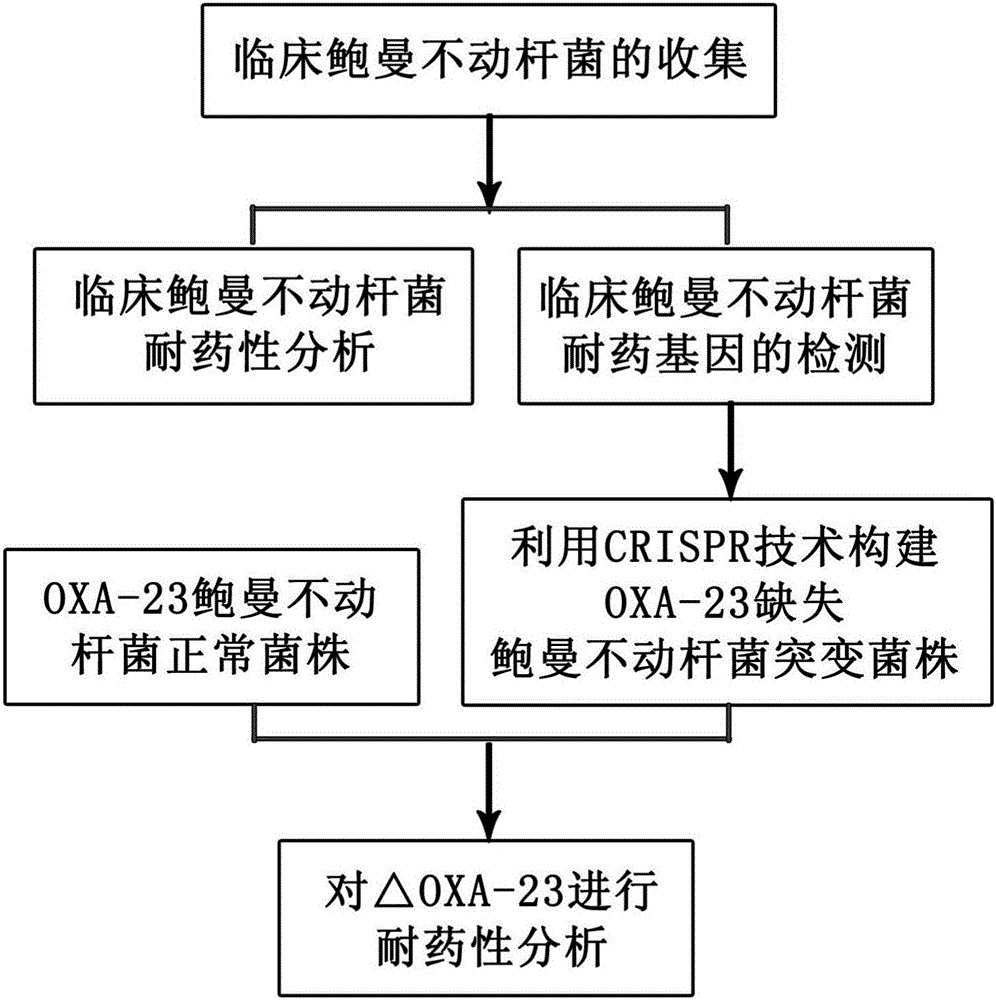
[0039] A method for eliminating drug resistance genes of Acinetobacter baumannii by using CRISPR-Cas9, the method is specifically:
[0040] 1. Collection of strains and plasmids used
[0041] Collected 106 non-repetitive imipenem-resistant Acinetobacter baumannii strains from clinical isolation, and 102 imipenem-sensitive strains. Experimental confirmation.
[0042] PET41a, pgRNA (44251), pwtCas9 (44250)
[0043] 2. Polymerase Chain Amplification (PCR)
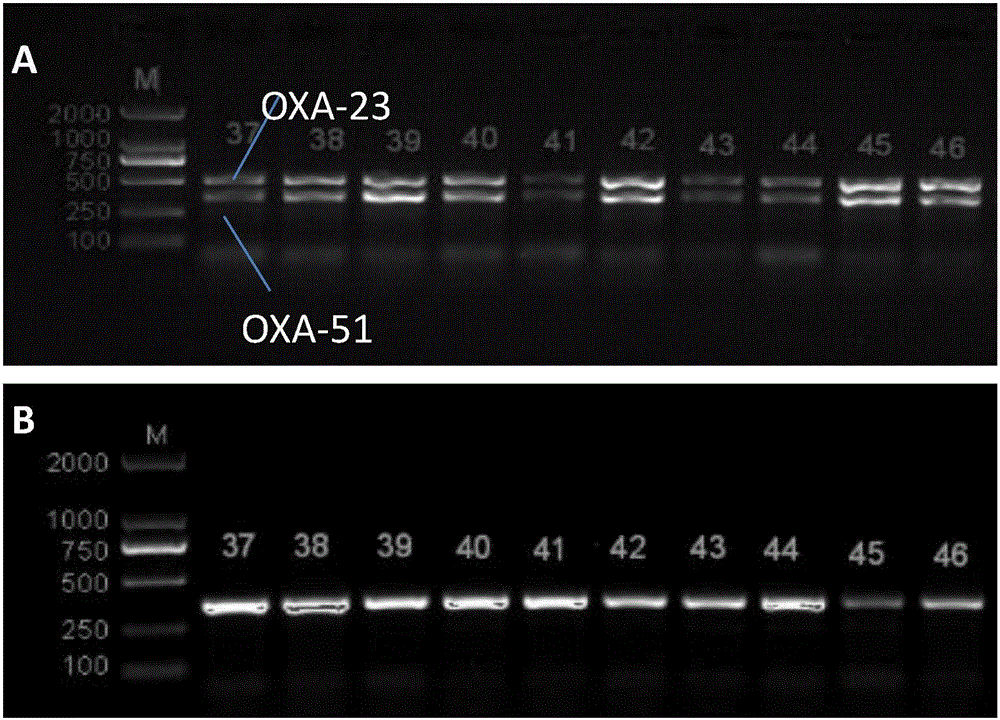
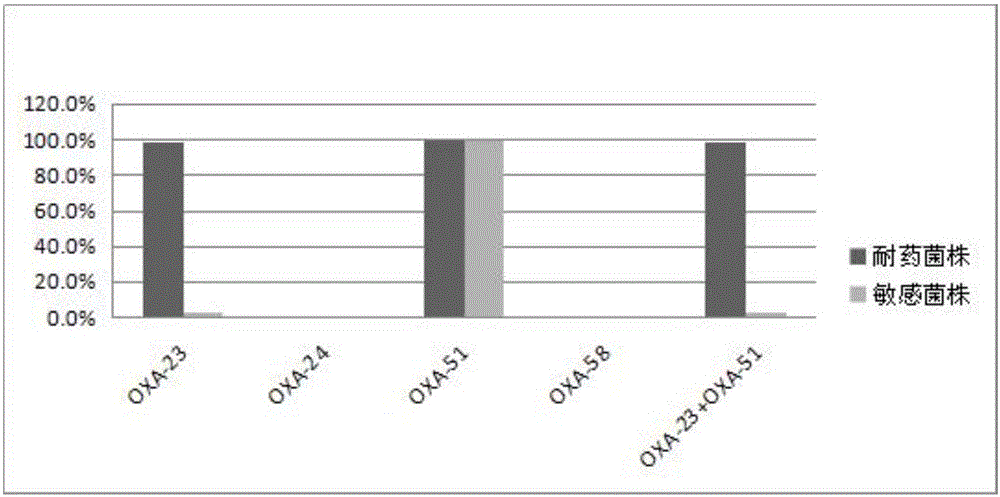
[0044]Bacterial DNA was extracted using the boiling method. Add 500 μL of the bacterial solution shaken overnight to a sterile EP tube, cook at 100°C for 10 minutes, and centrifuge at 13,000×g for 10 minutes. The obtained supernatant is the DNA template. For OXA-23, OXA-24, OXA-51, and OXA-58, multiplex PCR was used, and PCR technology was used for AdeABC and CarO gene detection. The primers used are shown in Table 1. The multiplex PCR reaction system (50 μL) includes 2 μL DNA template, 25 μL 2×TaqPCR MasterMix, 19 μL ddH2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com