SNP loci linked with blight resistant gene Fon-1 in watermelon, and markers thereof
A technology of resistance to fusarium wilt and fon-1, applied in DNA preparation, recombinant DNA technology, DNA/RNA fragments, etc., can solve the problems of difficulty in determining the phenotype of fusarium wilt at the seedling stage of watermelon, affecting positioning and acquisition, and shortening the breeding cycle , the effect of shortening the time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
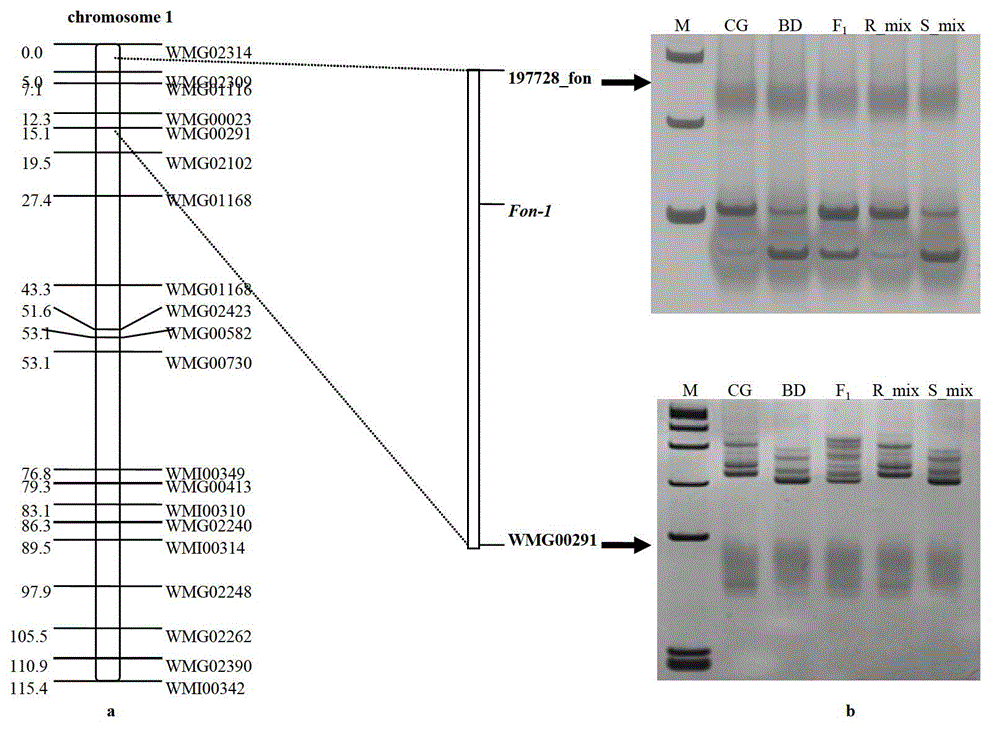
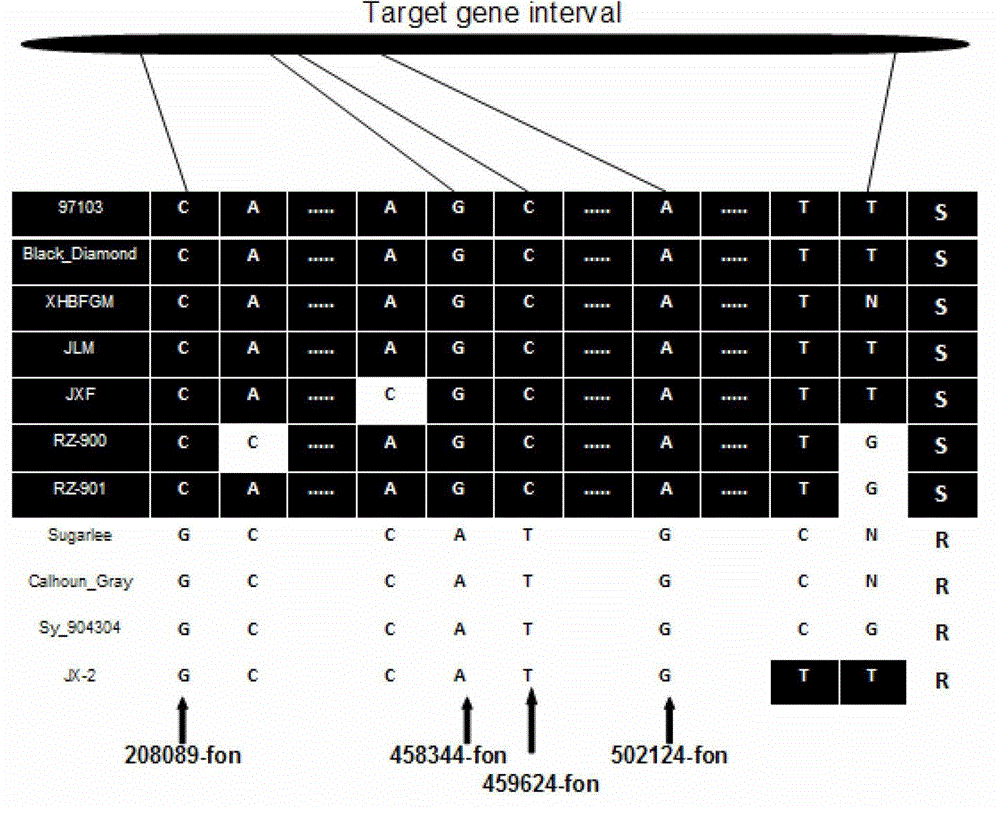
[0039] This example is the SNP sites and markers linked to the watermelon fusarium wilt resistance gene Fon-1 and the method for obtaining them.
[0040] The SNP sites and markers linked to the watermelon fusarium wilt resistance gene Fon-1 have the nucleotide base sequences of SEQ ID NO: 1-8 in the sequence list.
[0041] In the sequence listing, SEQ ID NO: 1 is the nucleotide base sequence specifically amplified by the primer 208089_fon for disease-resistant materials;
[0042] In the sequence listing, SEQ ID NO: 2 is the nucleotide base sequence specifically amplified by the primer 208089_fon to the susceptible material;
[0043] In the sequence listing, SEQ ID NO:3 is the nucleotide base sequence specifically amplified by the primer 458344_fon against disease-resistant materials;
[0044] In the sequence listing, SEQ ID NO:4 is the nucleotide base sequence specifically amplified by the primer 458344_fon to the susceptible material;
[0045] In the sequence listing, SEQ ID ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com