Recombinant thermostable dna polymerase and its application
A polymerase and heat-resistant technology, applied in the direction of enzymes, transferases, biochemical equipment and methods, etc., to achieve the effects of enhanced progression, long read length, and fast extension speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
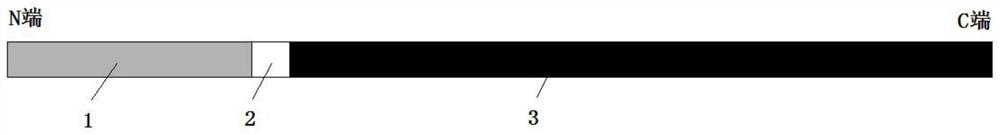
[0026] like figure 1 As shown, a recombinant thermostable DNA polymerase, including DNA binding domain 1, protein domain linker (protein linker) 2 and thermostable DNA polymerase 3 without 3'-5' exonuclease activity, DNA binding domain 1 is connected to the N-terminal of thermostable DNA polymerase 3 without 3'-5' exonuclease activity through linker 2.
[0027] In this embodiment, the DNA binding domain 1 is the Sso7d protein domain, and its amino acid sequence is shown in SEQ ID NO.1. The protein linker 2 has an amino acid sequence of Gly-Gly-Gly-Thr-Val, and the nucleotide sequence encoding the linker is shown in SEQ ID NO.2. The role of protein linker 2 is to connect two independent proteins or protein domains, while maintaining the independent activity and function of the two connected proteins or protein domains.
[0028] In this example, the thermostable DNA polymerase 3 without 3'-5' exonuclease activity is a Taq DNA polymerase deletion mutant, whose N-terminus has 28...
Embodiment 2
[0045] A recombinant thermostable DNA polymerase, including HMf protein, HMf protein is connected to Taq Δ289 DNA polymerase through protein linker, wherein, protein linker 2 is Gly-Gly-Gly-Thr-Val amino acid sequence, Taq Δ289 DNA The N-terminal of the polymerase lacks 289 amino acids, and contains two mutation points, R660D and F667Y.
[0046] The preparation method of the above recombinant heat-resistant DNA polymerase is as follows:
[0047] (1) Expression plasmid construction and expression strain transformation:
[0048] The Hmf protein coding sequence is artificially synthesized, and the amino acid sequence is shown in SEQ ID NO.7. And it was ligated into pET28a(sso7d-taqΔ289) by restriction endonuclease NcoI / SpeI to construct expression plasmid pET28a(hmf-taqΔ289). The target gene sequence is shown in SEQ ID NO.9. And transformed into Escherichia coli expression strain BL21 (DE3).
[0049] (2) Induced expression and purification of Hmf-TaqΔ289 polymerase
[0050] T...
Embodiment 3
[0052] A recombinant heat-resistant DNA polymerase, including Sso7d protein domain, the Sso7d protein domain is connected to a Tth polymerase mutant without 3`-5`exonuclease activity through a protein linker, wherein, the protein linker 2 is The Gly-Gly-Gly-Thr-Val amino acid sequence, the amino acid sequence of the Tth polymerase mutant is shown in SEQ ID NO.4, and the target gene sequence is shown in SEQ ID NO.10.
[0053] The preparation method of the above recombinant heat-resistant DNA polymerase is as follows:
[0054] (1). Expression plasmid construction and expression strain transformation:
[0055] The DNA sequence encoding TthΔ289 polymerase was cloned from Thermus thermophilus HB8 genomic DNA (purchased from TAKAArgA, 3071), and the gene sequence is shown in SEQ ID NO.4. The homologous sequence of TaqΔ289 was replaced by homologous recombination, and the expression plasmid pET28a(sso7d-tthΔ289) was constructed and transformed into E. coli expression strain BL21(DE3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com