Method for rapidly preparing Sanger sequencing template
A sequencing and template technology, applied in the field of bioengineering, achieves the effects of high sequencing success rate, overcoming high cost, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
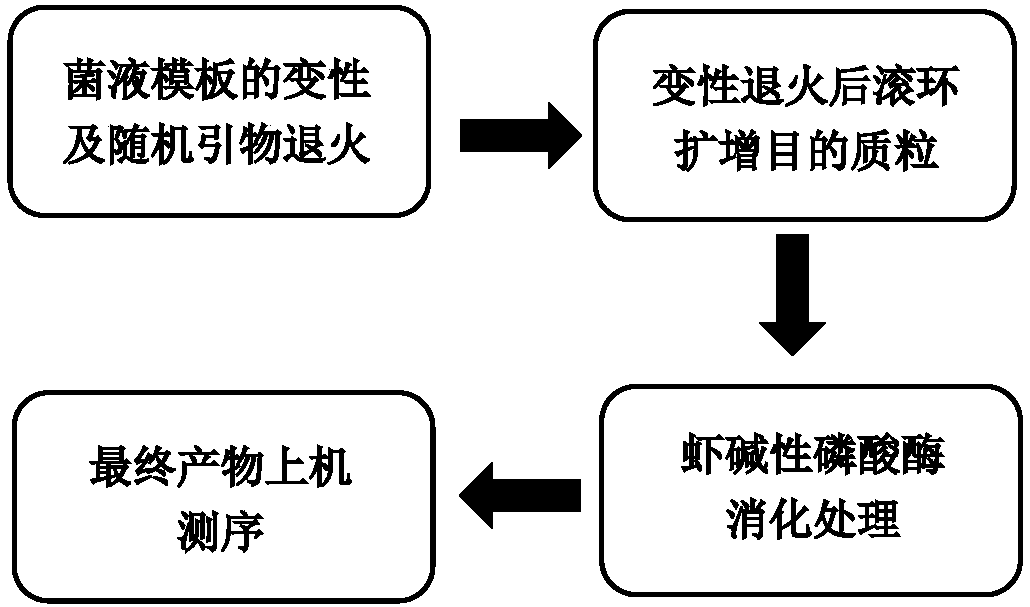
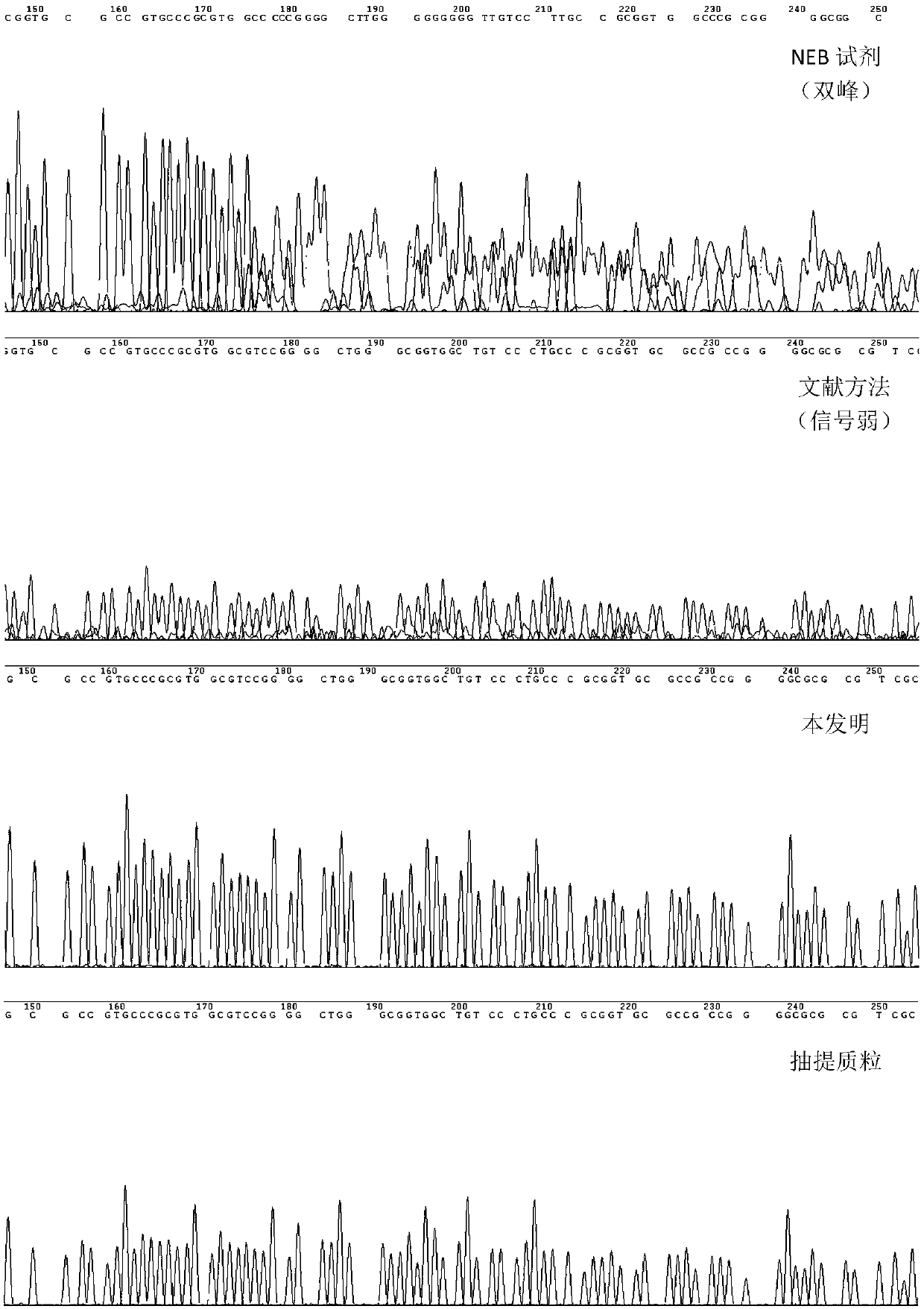
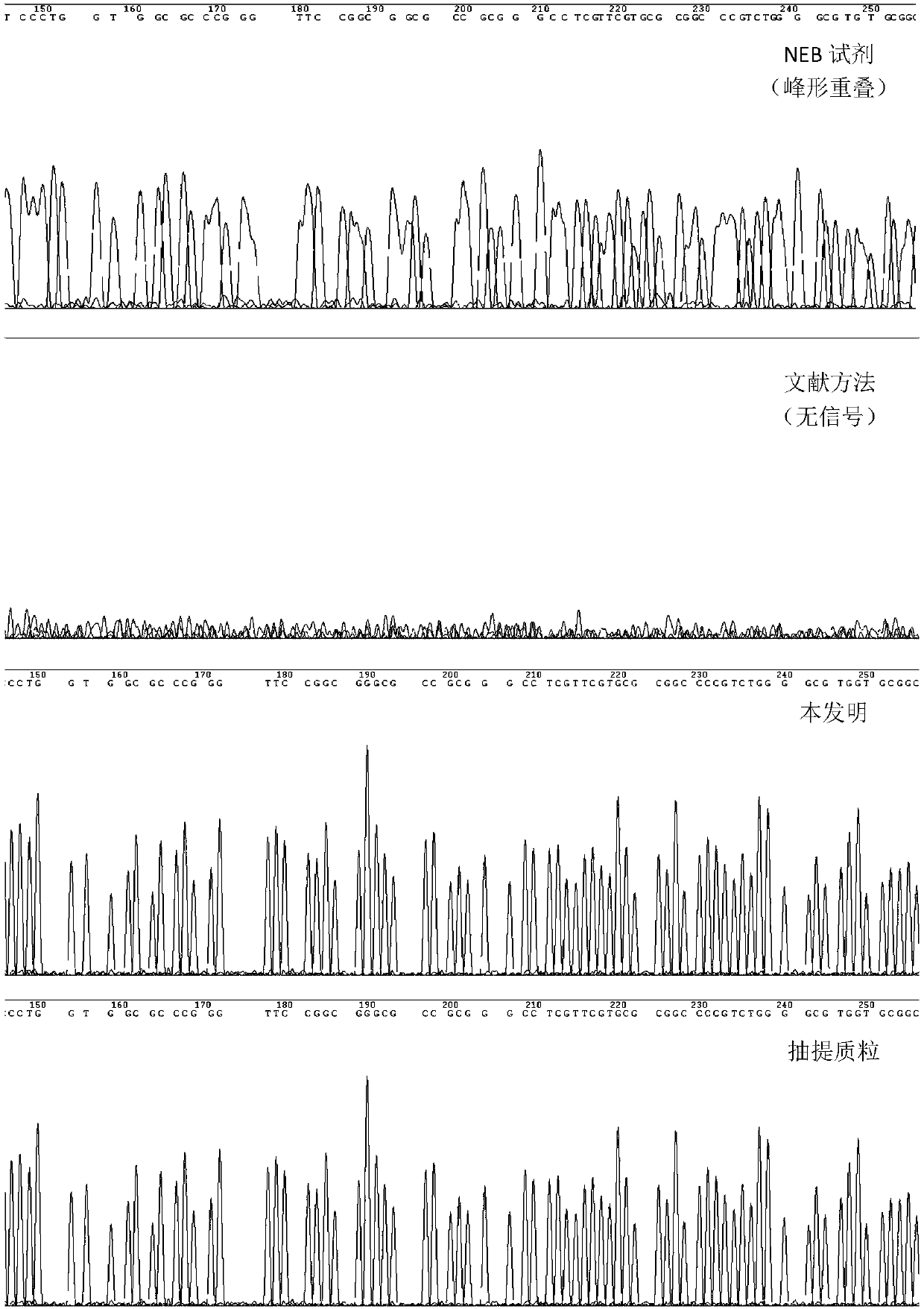
[0028] This example compares the sequencing effects of preparing Sanger sequencing templates within four hours using three different rolling circle amplification methods for preparing sequencing templates:
[0029] A. NEB phi29DNA Polymerase Reagent (Product No. M0269S, this reagent provides some supporting reagents and experimental operation scheme for RCA reaction) (hereinafter referred to as "NEB Reagent (phi29DNA Polymerase)")
[0030]B. Cited from literature (Frank B. Dean., John R. Nelson., Theresa L. Giesler., and Roger S. Lasken. 2001. Rapid Amplification of Plasmid and Phage DNA Using Phi29DNA Polymerase and Multiply-Primed Rolling Circle Amplification. Genome Research. 1095-1099) of the RCA protocol (indicated below as "methods in the literature")
[0031] C. The optimized RCA scheme provided by this application (hereinafter referred to as "this application")
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com