PET series carrier forward sequencing primers and sequencing method
A technology of forward sequencing and primer sequence, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, DNA/RNA fragments, etc. Experimental cycle, high success rate, and the effect of increased success rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
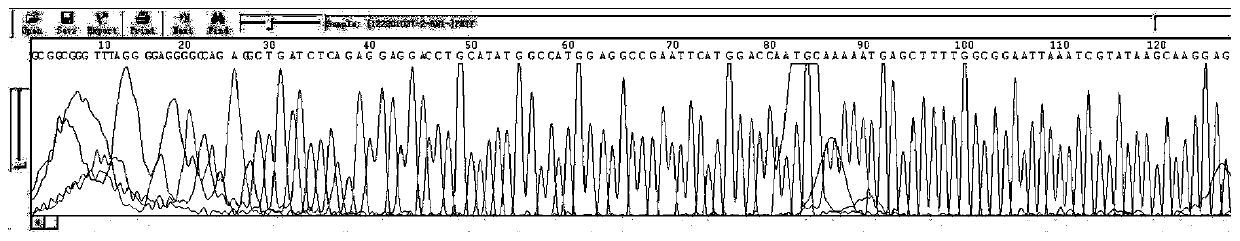
[0025] In this example, the PET series vector containing the target gene was sequenced, wherein the forward sequencing primer used was 5'-GCGAAATTAATACGACTCACTAT-3', the length of the primer was 23bp, and the Tm temperature was 47.4°C.
[0026] The specific sequencing steps are as follows:
[0027] (1) Dilute the primers with ddH2O to a concentration of 5pmole / μl, and configure the following system in a 96-well PCR plate: 1 μl of PET carrier containing the target gene, 1 μl of forward sequencing primer, 2 μl of BigDye mix, ddH 2 O 1 μl;
[0028] (2) Seal the PCR plate with a silica gel pad, put it into a centrifuge and centrifuge it out for a short time, and run it in the PCR instrument according to the following procedures: the PCR program is 96°C for 1min; 96°C for 10s, 50°C for 5s, 60°C for 4min, 25 cycles Cycle; keep warm at 4°C;
[0029] (3) After PCR, take out the 96PCR plate, add 2μl EDTA / NaAC and 20ul cold alcohol to each well, and let stand for 8min;
[0030] (4) S...
Embodiment 2
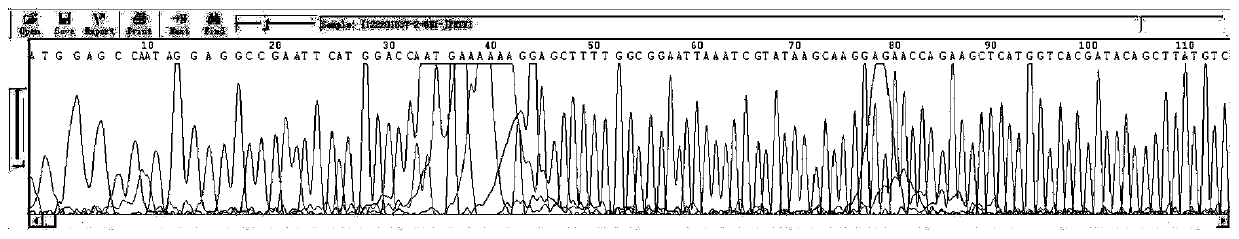
[0038] In this embodiment, the PET series vector of the target gene is sequenced, wherein the forward sequencing primer used is 5'-RAWWKTAATACGACTCACTAT-3', the length of the primer is 21 bp, and the Tm temperature is 39.5°C.
[0039] The specific sequencing steps are as follows:
[0040] (1) Dilute the primers with ddH2O to a concentration of 3 pmole / μl, and configure the following system in a 96-well PCR plate: 1 μl of PET carrier of the target gene, 1 μl of forward sequencing primer, 2 μl of BigDyemix, ddH 2 O 1 μl;
[0041] (2) Seal the PCR plate with a silica gel pad, put it into a centrifuge and centrifuge it out for a short time, and run it in the PCR instrument according to the following procedures: the PCR program is 96°C for 1min; 96°C for 10s, 50°C for 5s, 60°C for 4min, 25 cycles Cycle; keep warm at 4°C;
[0042] (3) After PCR, take out the 96PCR plate, add 1μl EDTA / NaAC and 12ul cold alcohol to each well, and let stand for 8min;
[0043] (4) Shake gently on a v...
Embodiment 3
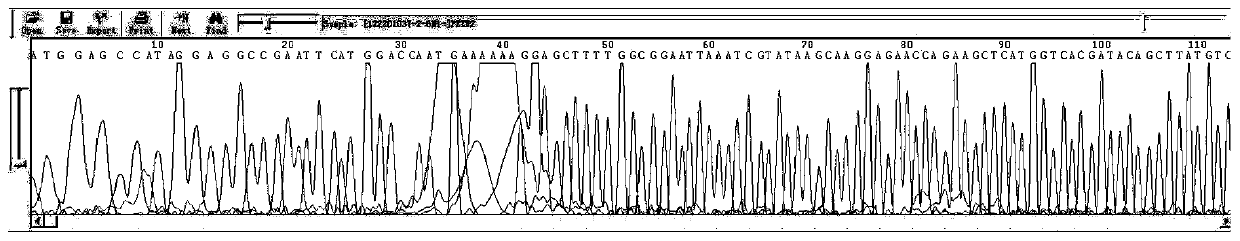
[0051] In this example, the PET serial vector of the target gene was sequenced, wherein the forward sequencing primer used was 5'-RAWWKTAATACGACTCACTA-3', the length of the primer was 20 bp, and the Tm temperature was 38.5°C.
[0052] The specific sequencing steps are as follows:
[0053](1) Dilute the primers with ddH2O to a concentration of 7pmole / μl, and configure the following system in a 96-well PCR plate: 1 μl of PET carrier containing the target gene, 1 μl of forward sequencing primer, 2 μl of BigDye mix, ddH 2 O 1 μl;
[0054] (2) Seal the PCR plate with a silica gel pad, put it into a centrifuge and centrifuge it out for a short time, and run it in the PCR instrument according to the following procedures: the PCR program is 96°C for 1min; 96°C for 10s, 50°C for 5s, 60°C for 4min, 25 cycles Cycle; keep warm at 4°C;
[0055] (3) After PCR, take out the 96PCR plate, add 3μl EDTA / NaAC and 24ul cold alcohol to each well, and let stand for 8min;
[0056] (4) Shake gently...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com