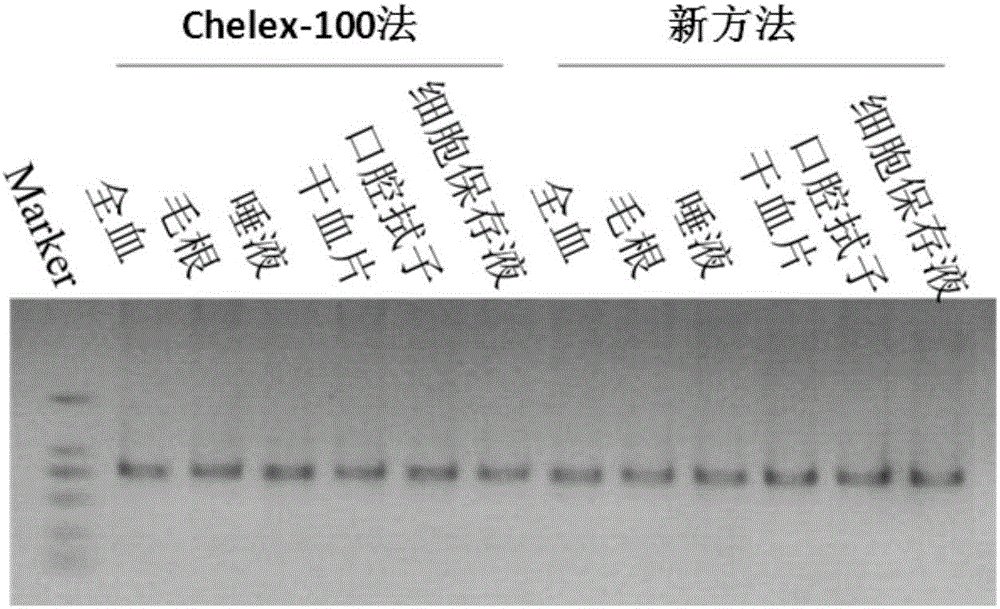
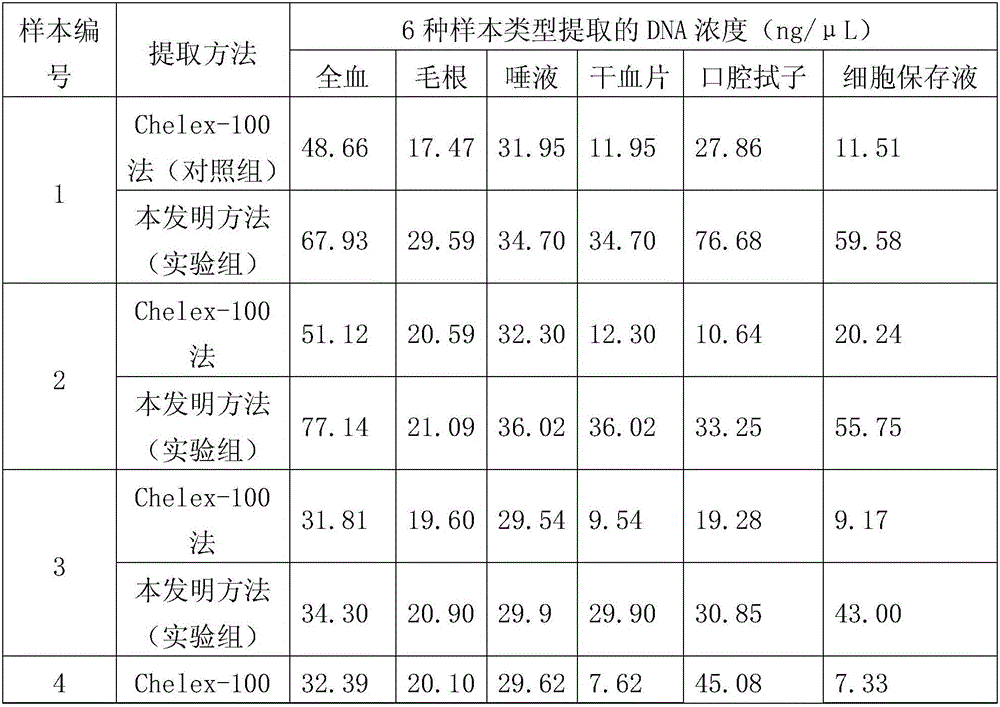
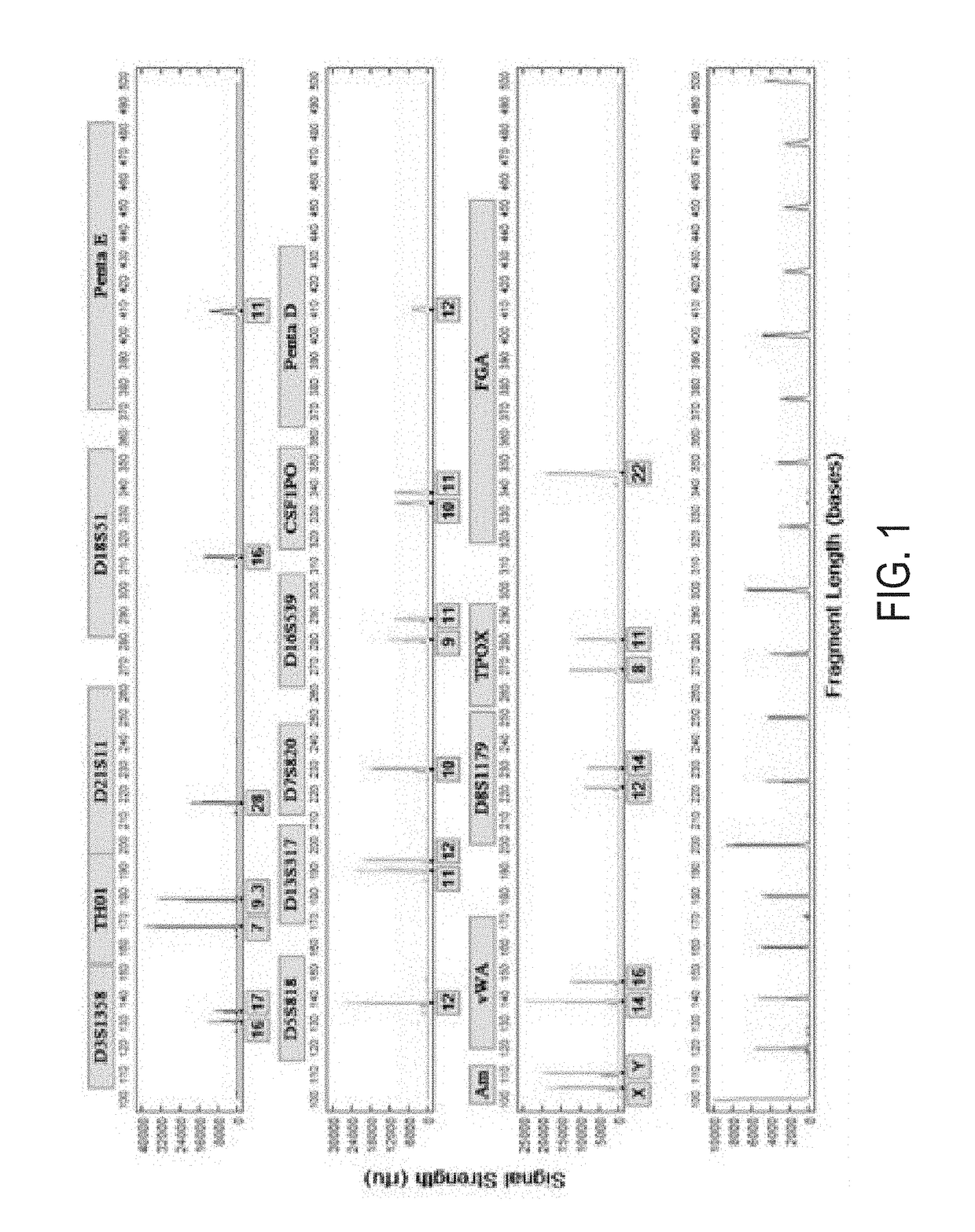
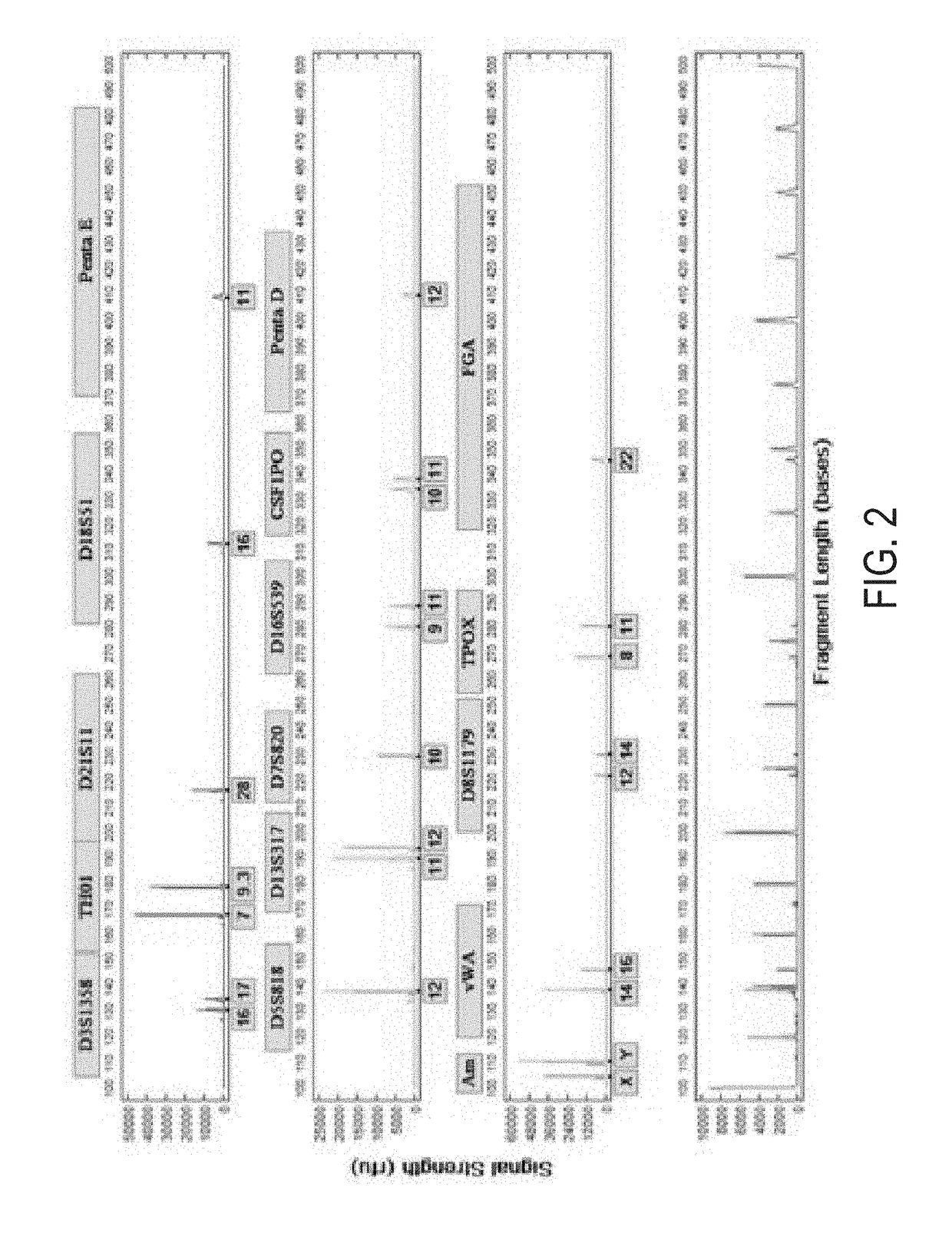
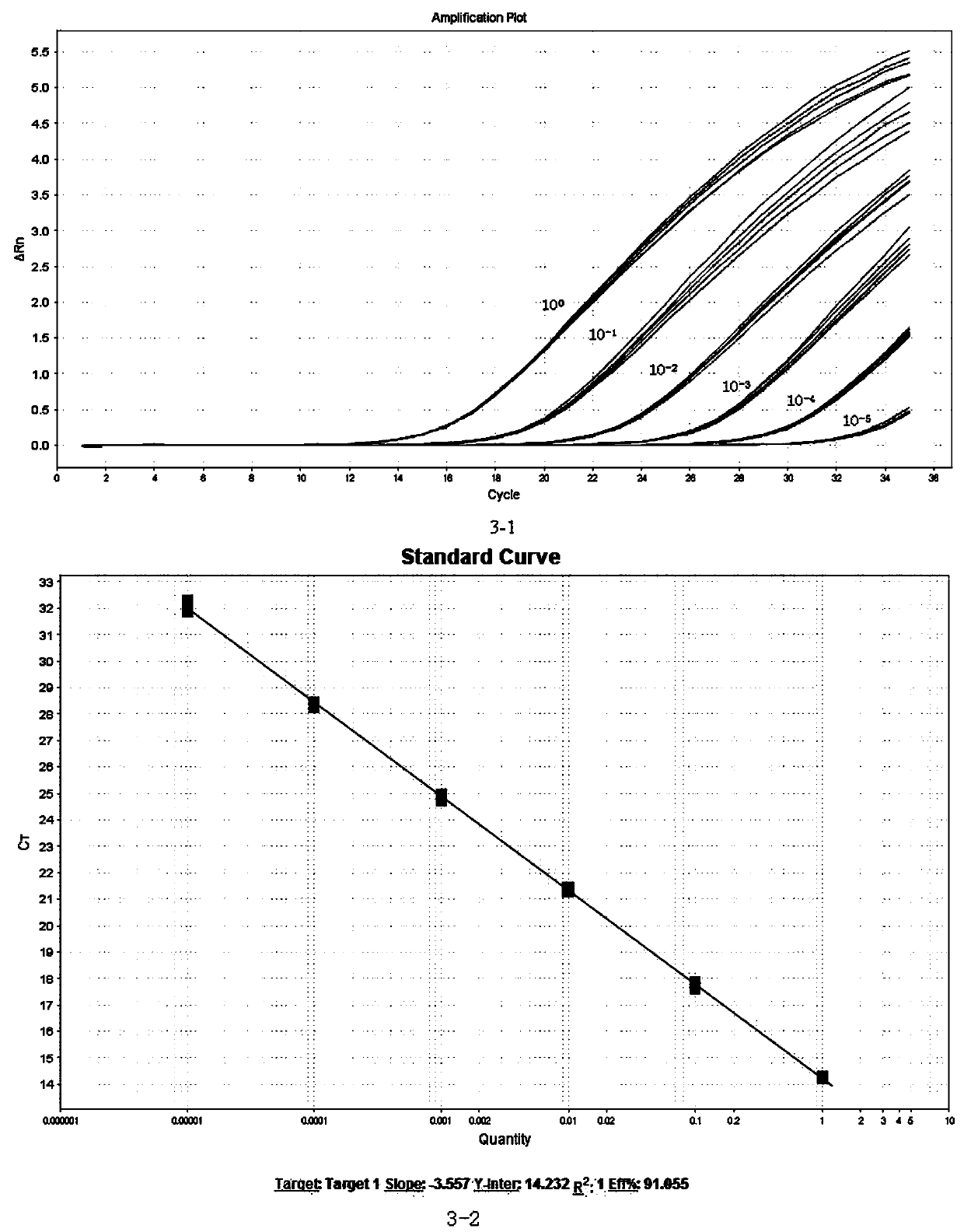
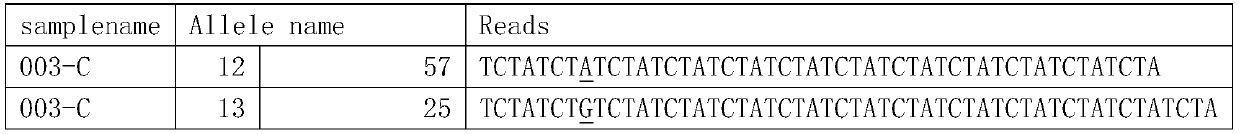
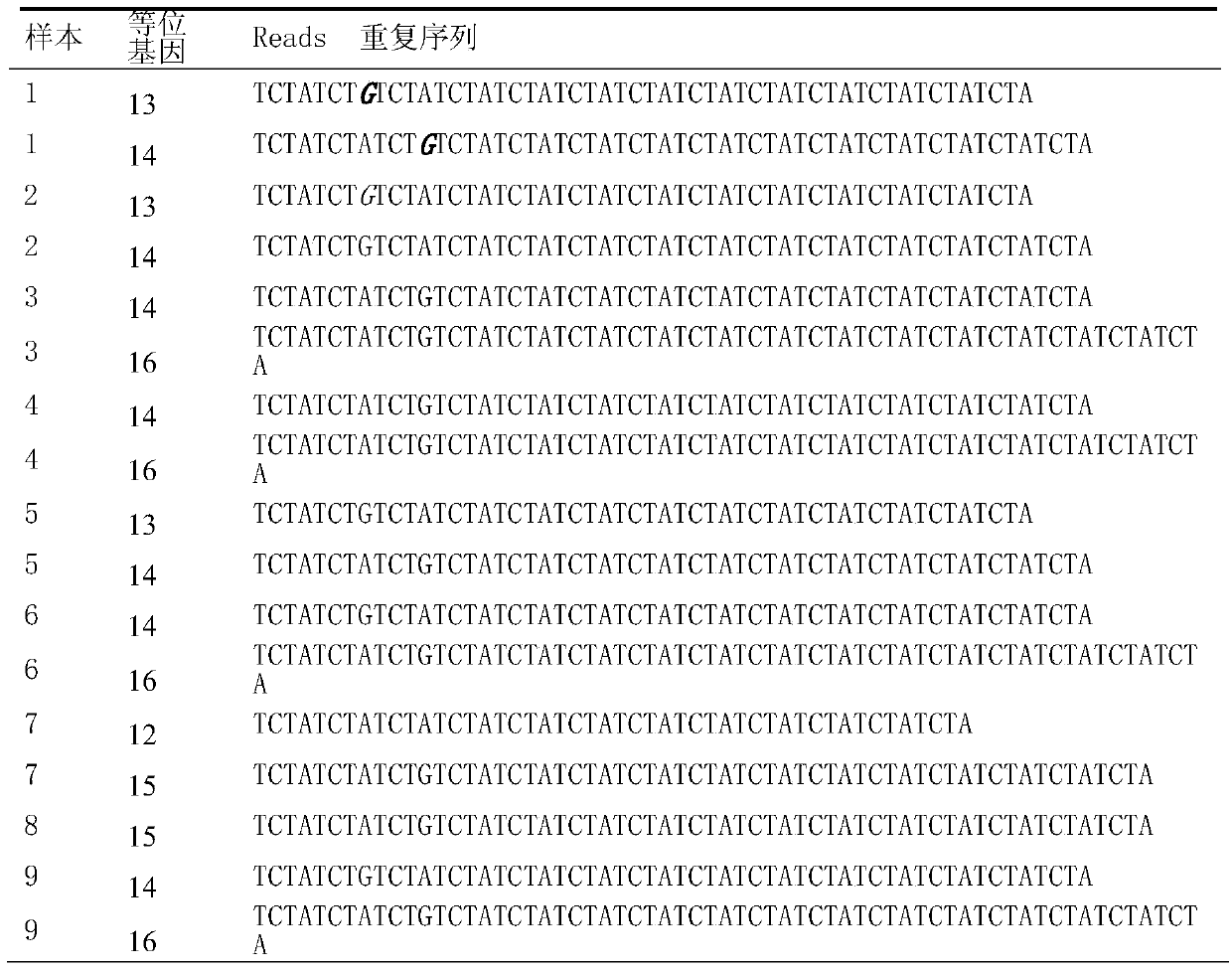
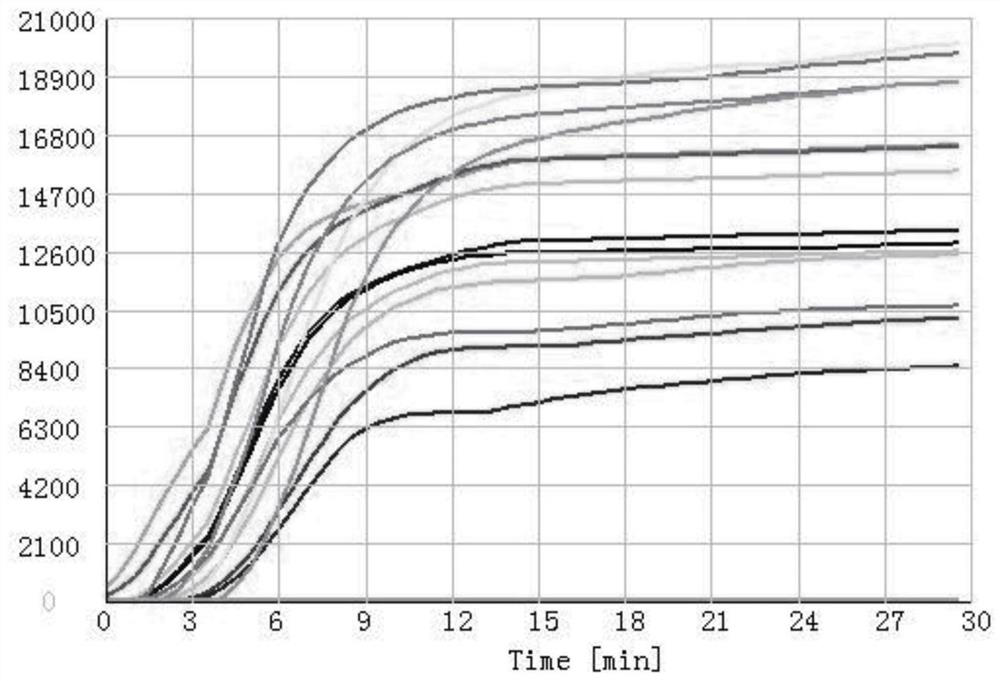
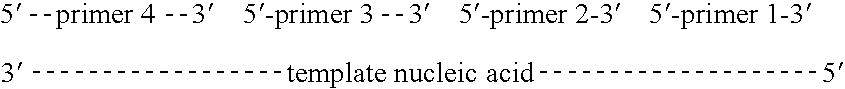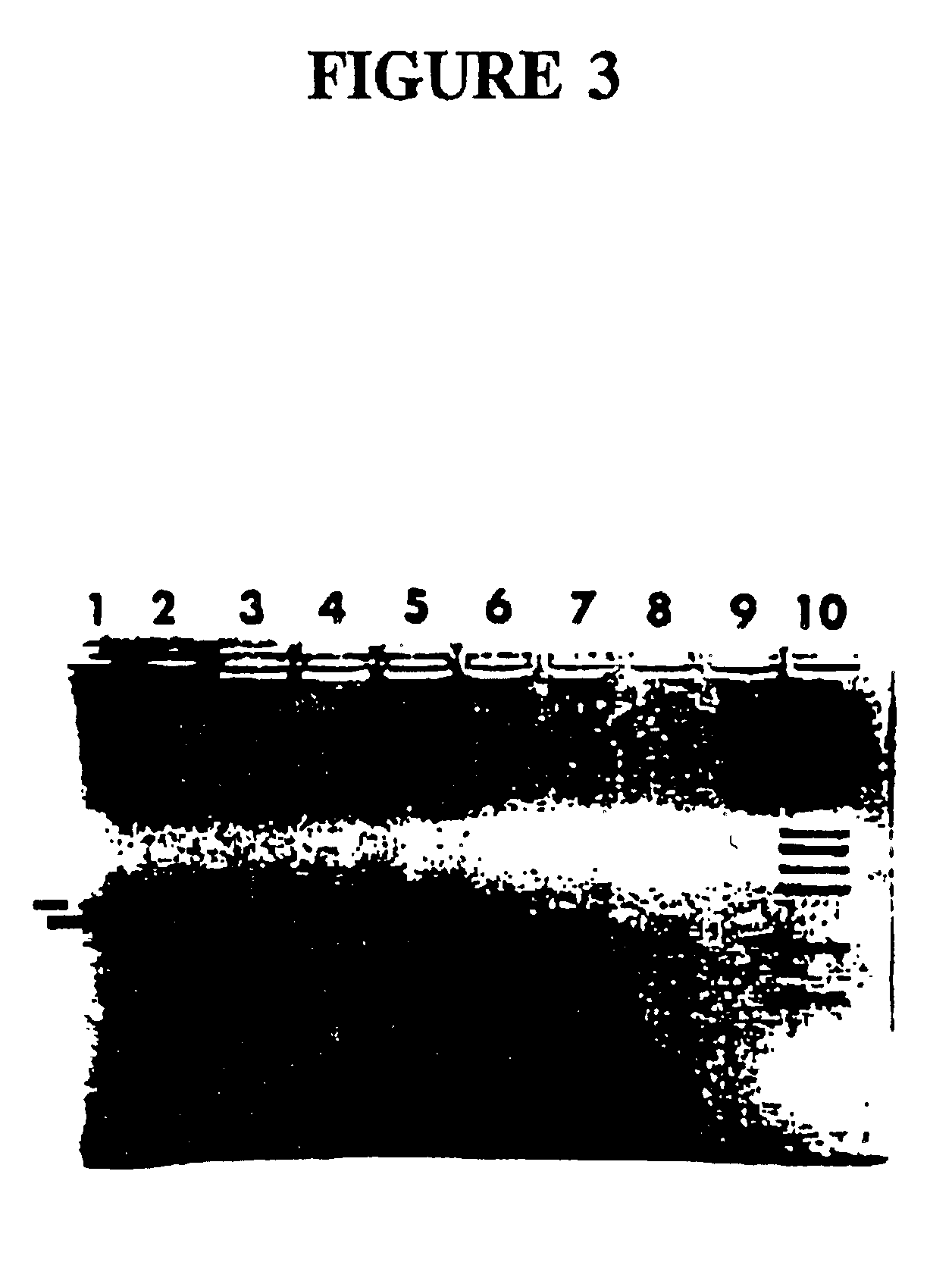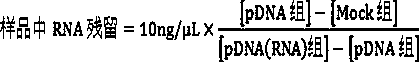Patents
Literature
53 results about "Rapid dna" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Methods of amplifying and sequencing nucleic acids
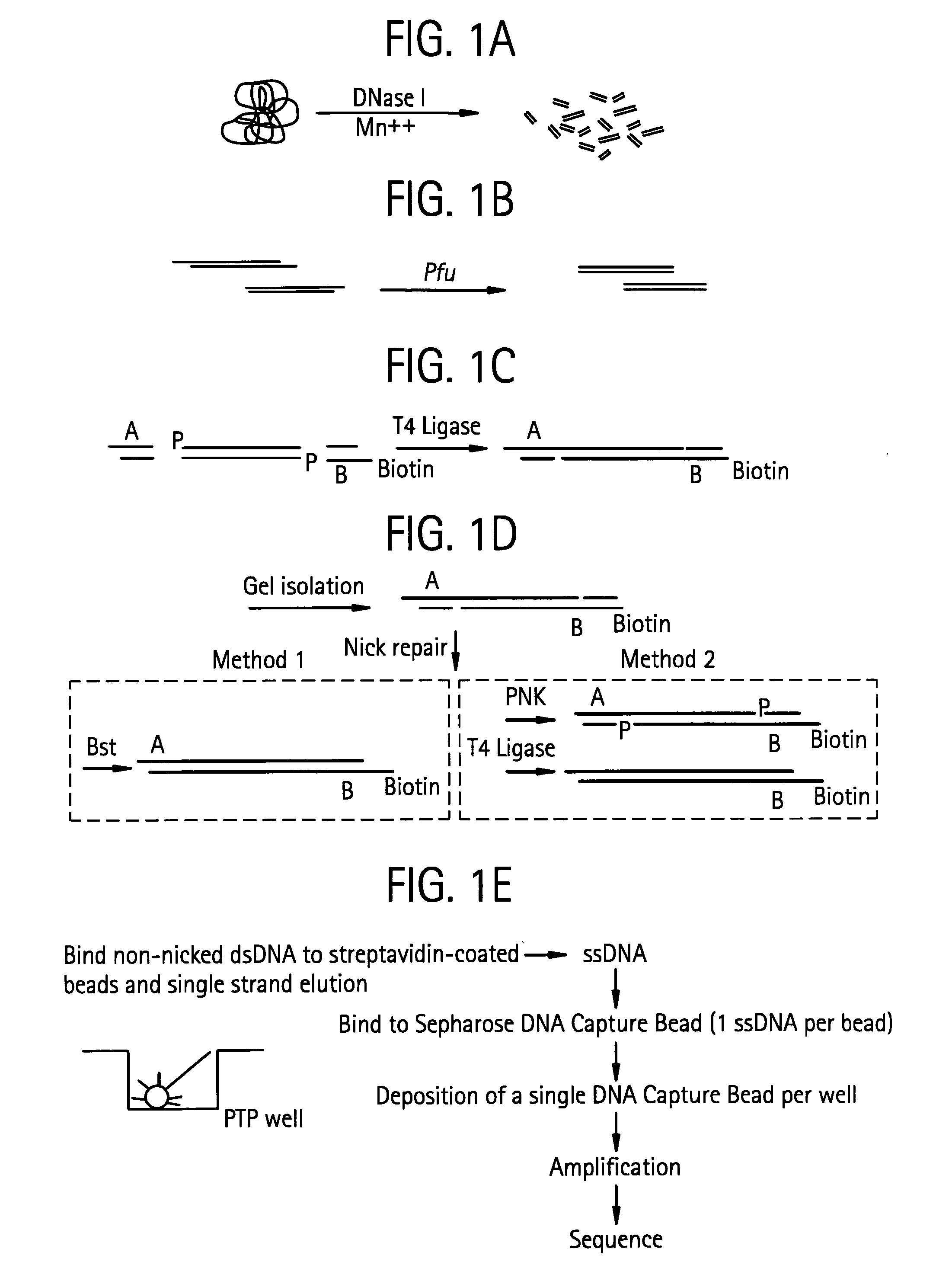
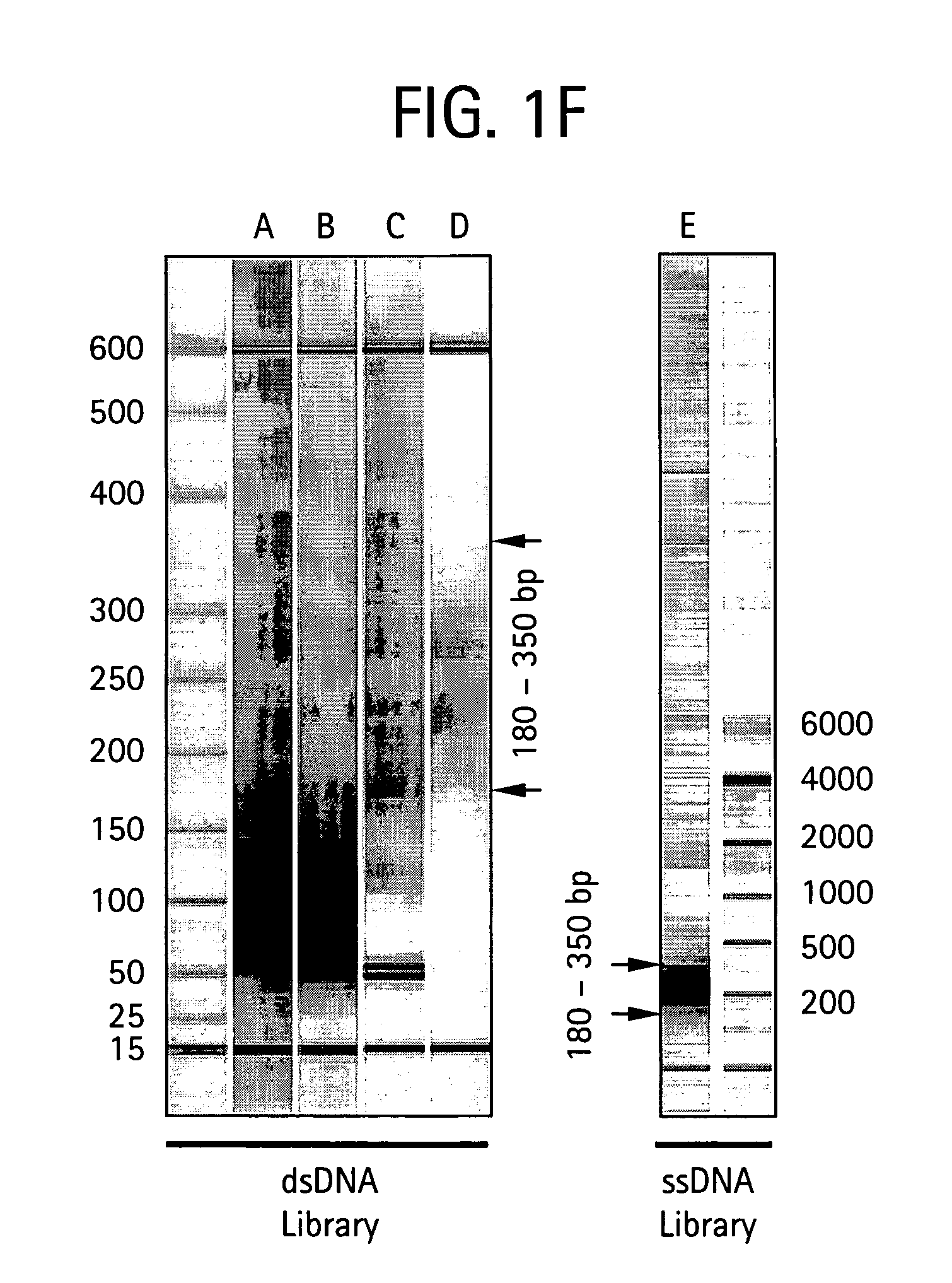
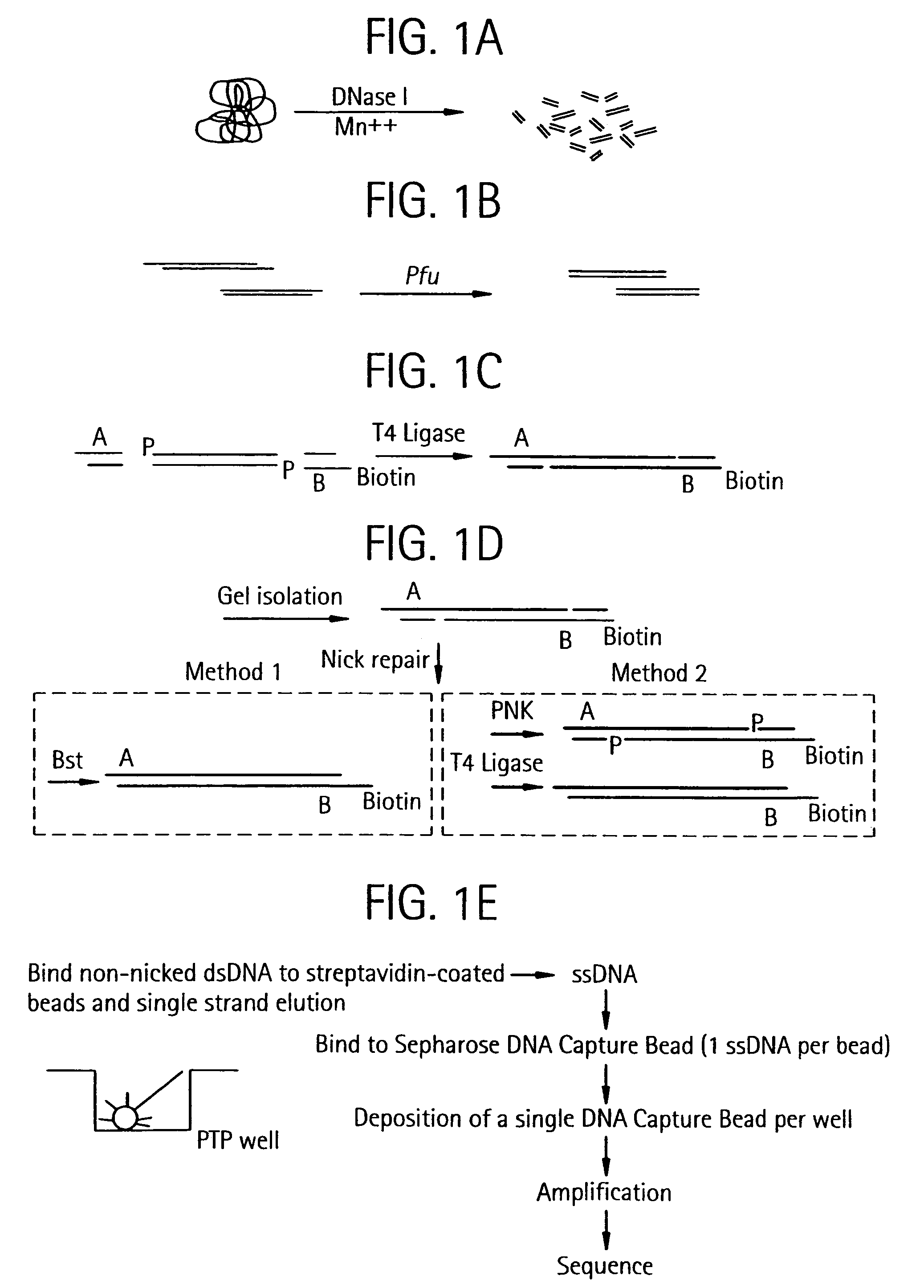
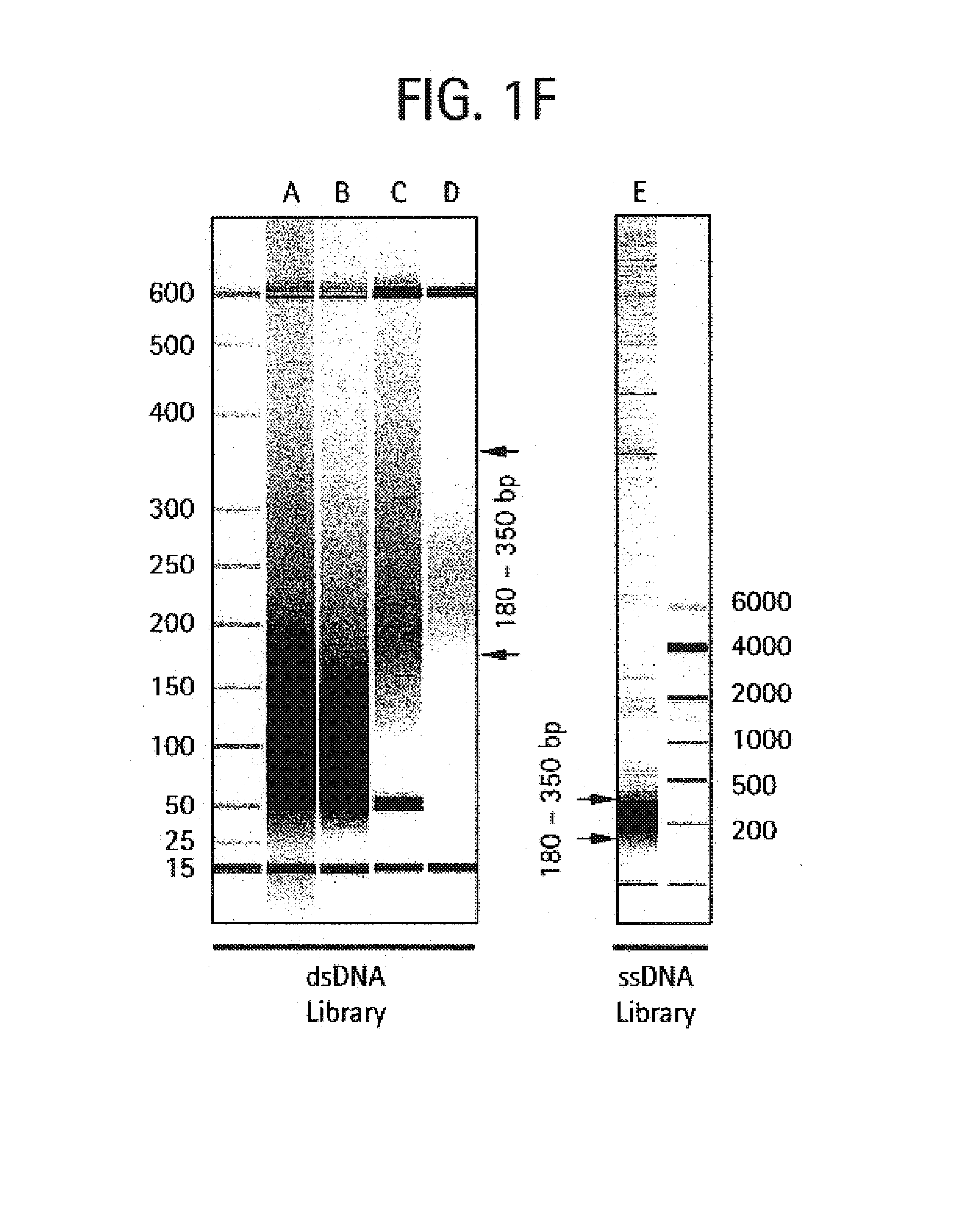
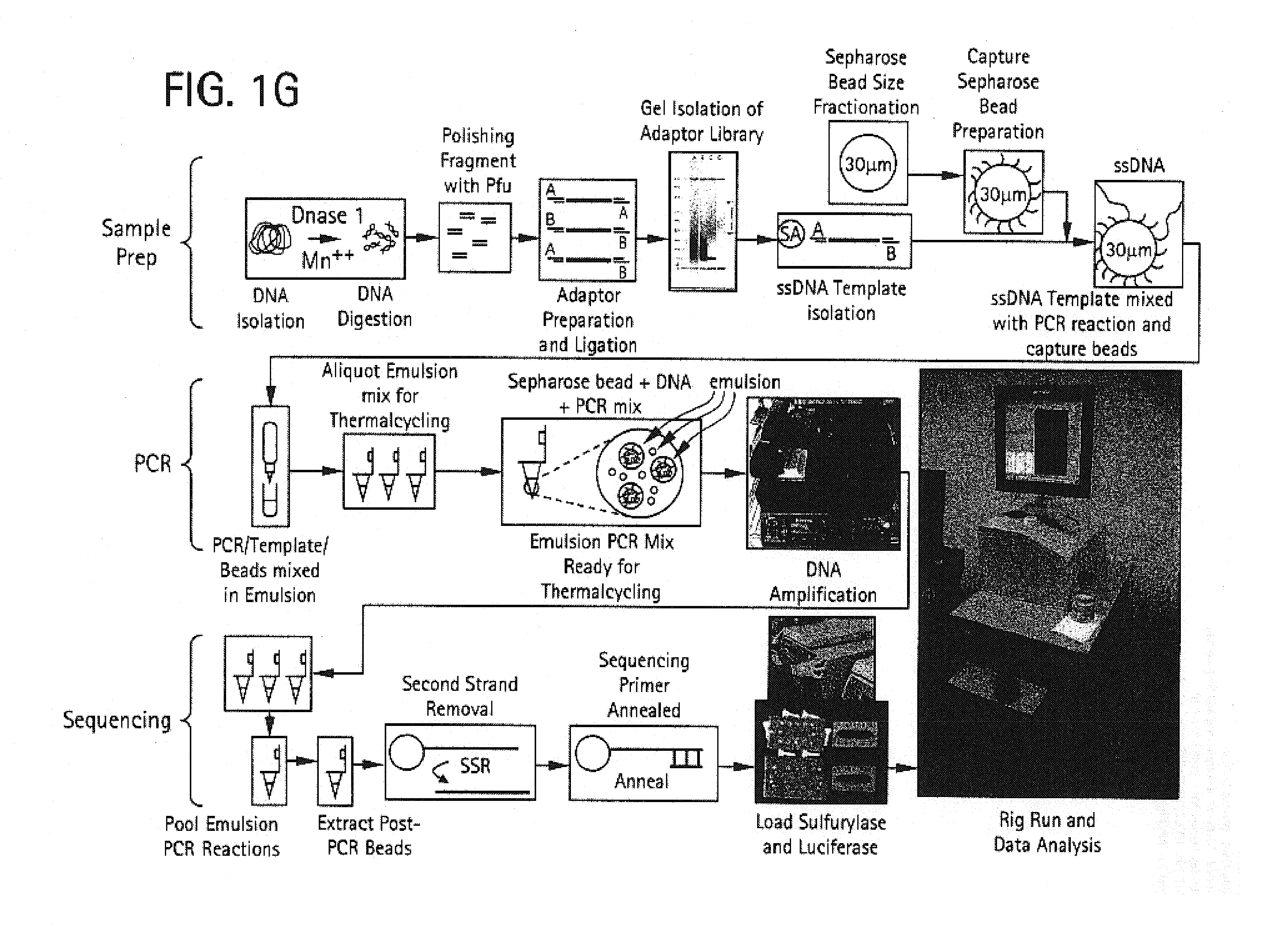
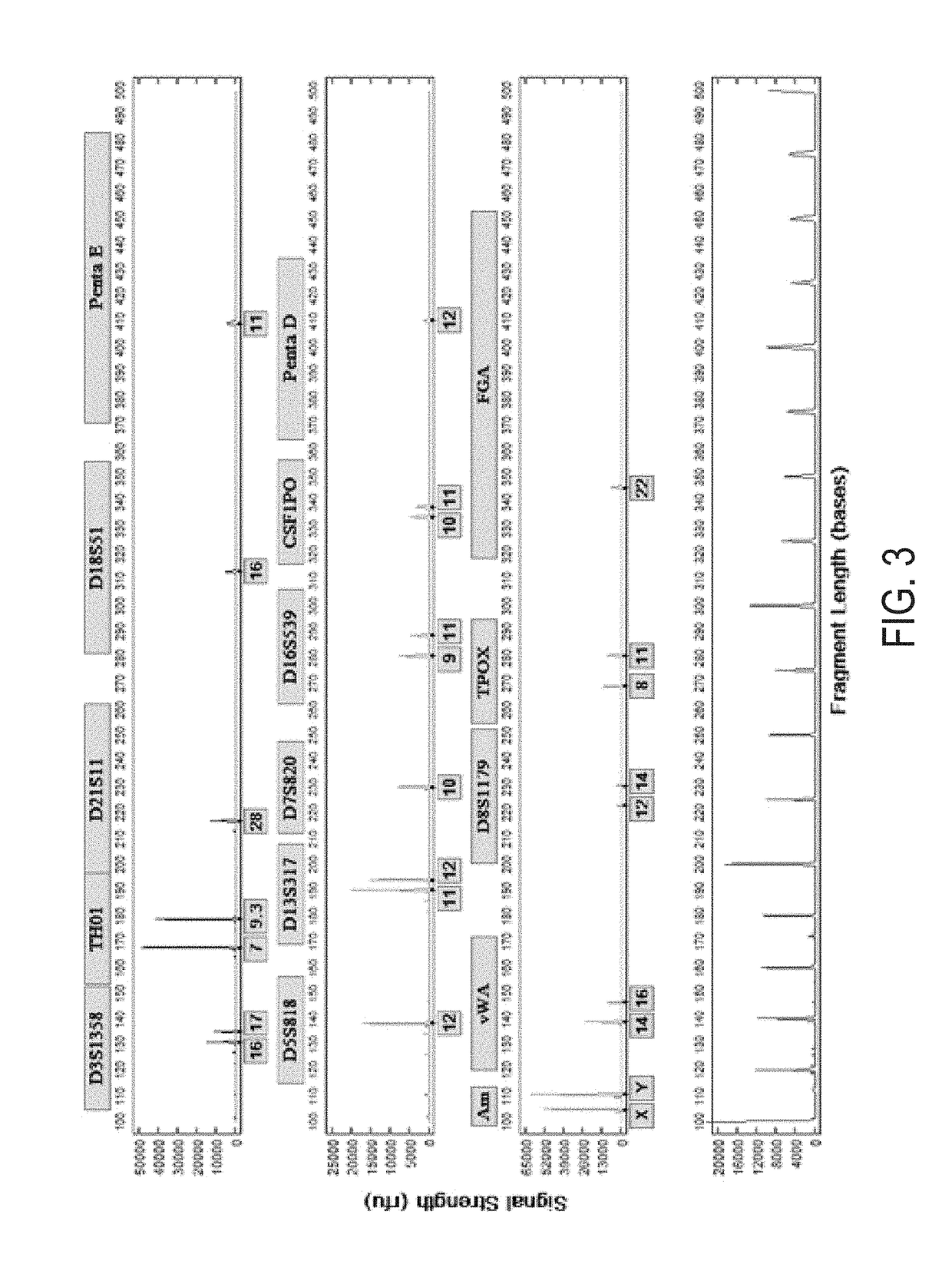
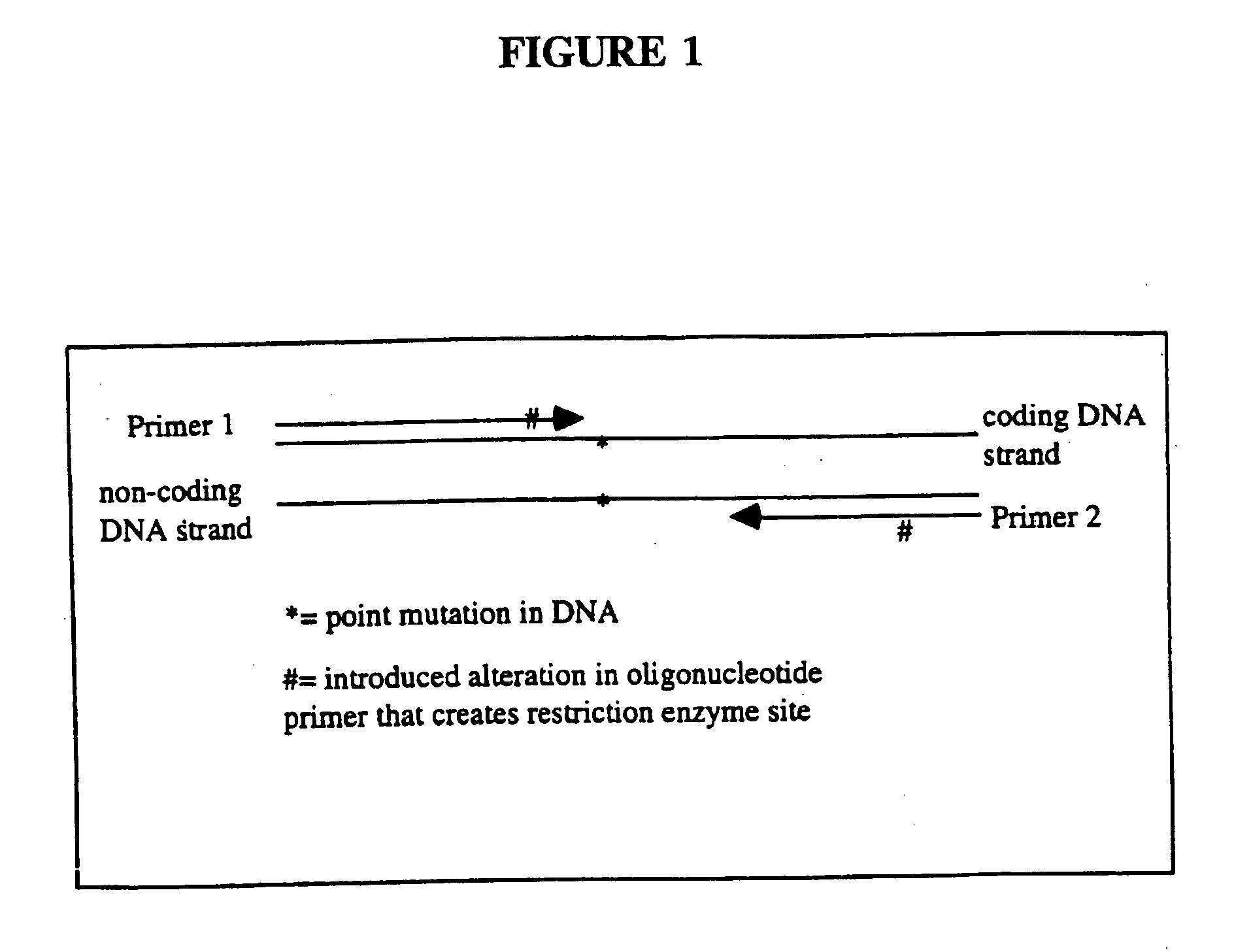
An apparatus and method for performing rapid DNA sequencing, such as genomic sequencing, is provided herein. The method includes the steps of preparing a sample DNA for genomic sequencing, amplifying the prepared DNA in a representative manner, and performing multiple sequencing reaction on the amplified DNA with only one primer hybridization step.
Owner:454 LIFE SCIENCES CORP
Methods of amplifying and sequencing nucleic acids
An apparatus and method for performing rapid DNA sequencing, such as genomic sequencing, is provided herein. The method includes the steps of preparing a sample DNA for genomic sequencing, amplifying the prepared DNA in a representative manner, and performing multiple sequencing reaction on the amplified DNA with only one primer hybridization step.
Owner:454 LIFE SCIENCES CORP
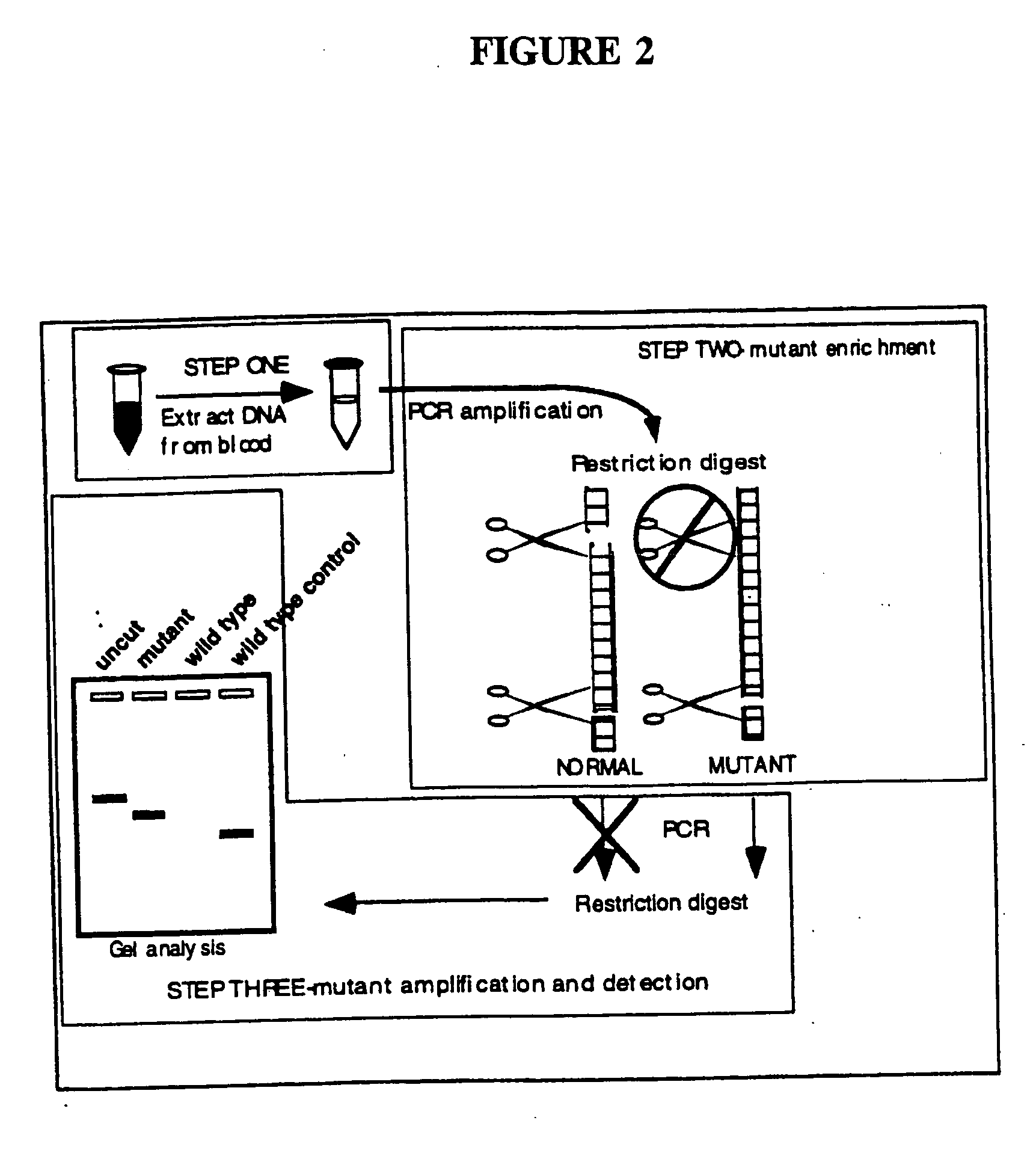
Detection of extracellular tumor-associated nucleic acid in blood plasma or serum using nucleic acid amplification assays
InactiveUS6939675B2Antibody mimetics/scaffoldsMicrobiological testing/measurementNeoplasmBlood plasma
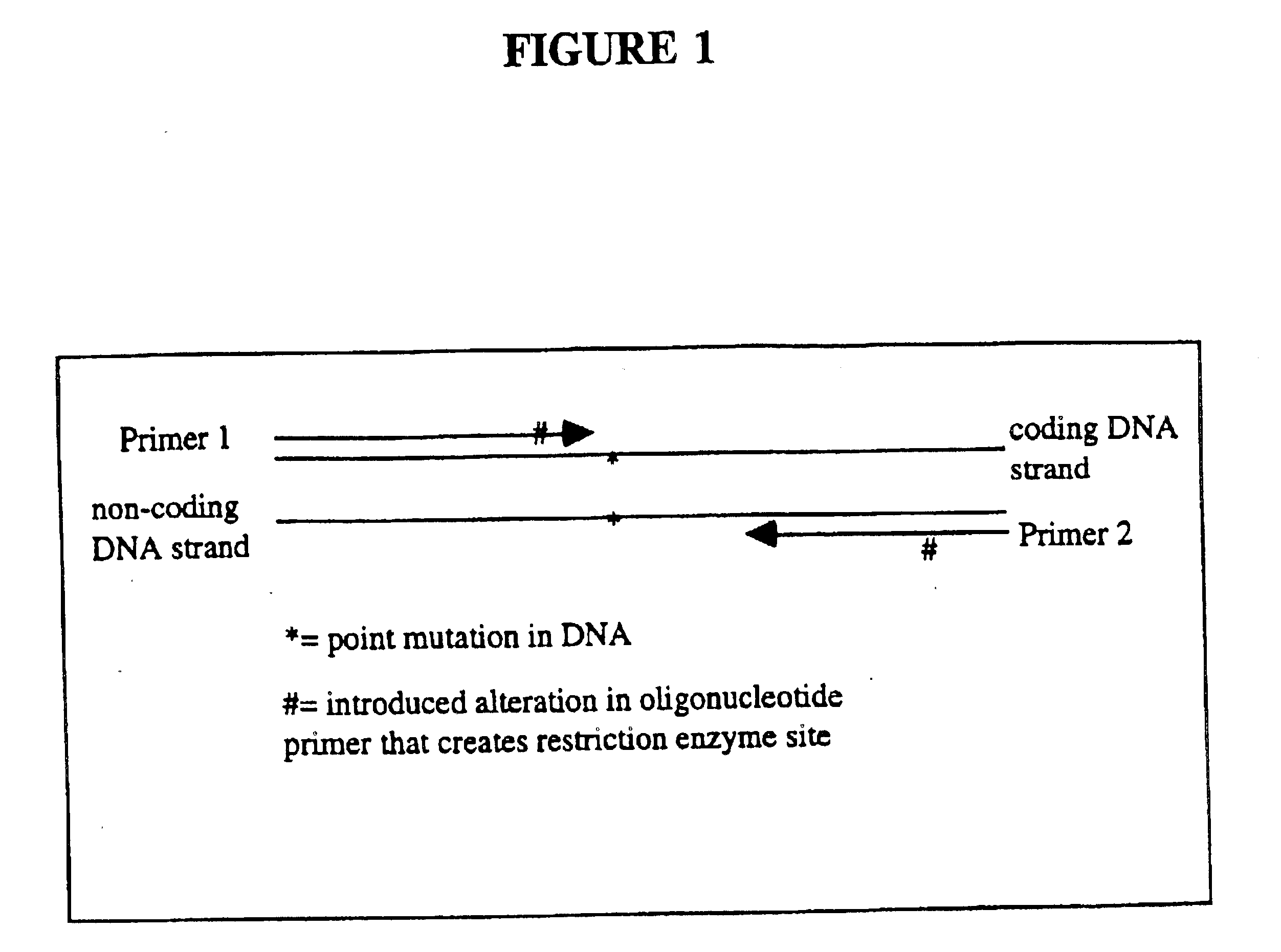
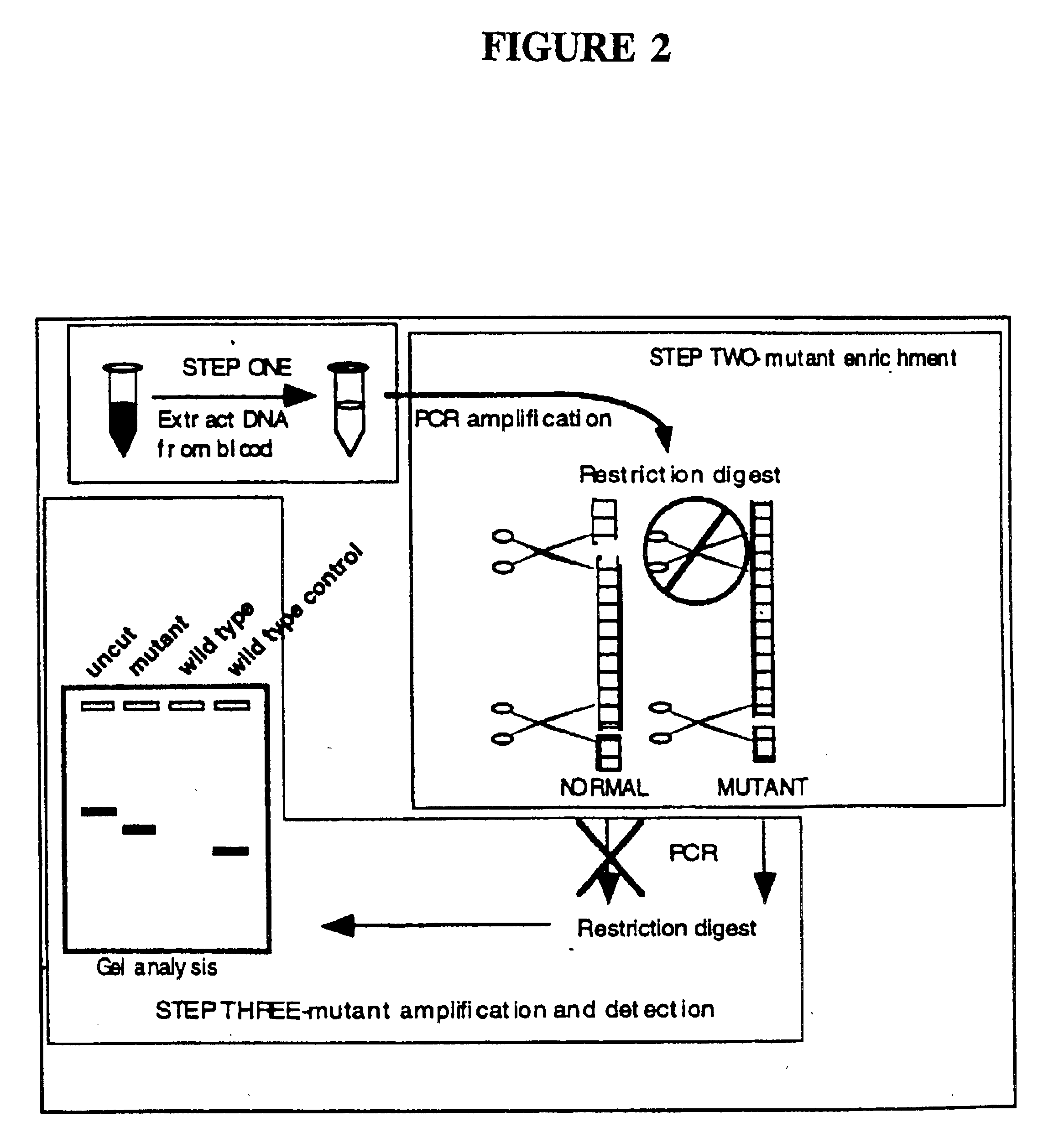
This invention relates to detection of specific extracellular nucleic acid in plasma or serum fractions of human or animal blood associated with neoplastic or proliferative disease. Specifically, the invention relates to detection of nucleic acid derived from mutant oncogenes or other tumor-associated DNA, and to those methods of detecting and monitoring extracellular mutant oncogenes or tumor-associated DNA found in the plasma or serum fraction of blood by using rapid DNA extraction followed by nucleic acid amplification with or without enrichment for mutant DNA. In particular, the invention relates to the detection, identification, or monitoring of the existence, progression or clinical status of benign, premalignant, or malignant neoplasms in humans or other animals that contain a mutation that is associated with the neoplasm through detection of the mutated nucleic acid of the neoplasm in plasma or serum fractions. The invention permits the detection of extracellular, tumor-associated nucleic acid in the serum or plasma of humans or other animals recognized as having a neoplastic or proliferative disease or in individuals without any prior history or diagnosis of neoplastic or proliferative disease. The invention provides the ability to detect extracellular nucleic acid derived from genetic sequences known to be associated with neoplasia, such as oncogenes, as well as genetic sequences previously unrecognized as being associated with neoplastic or proliferative disease. The invention thereby provides methods for early identification of colorectal, pancreatic, lung, breast, bladder, ovarian, lymphoma and all other malignancies carrying tumor-related mutations of DNA and methods for monitoring cancer and other neoplastic disorders in humans and other animals.
Owner:PENN STATE RES FOUND
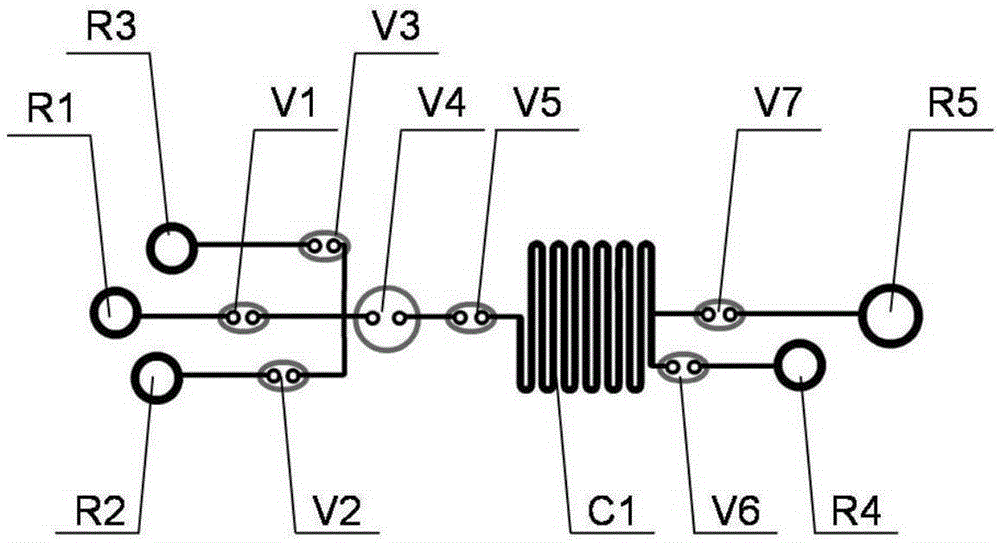
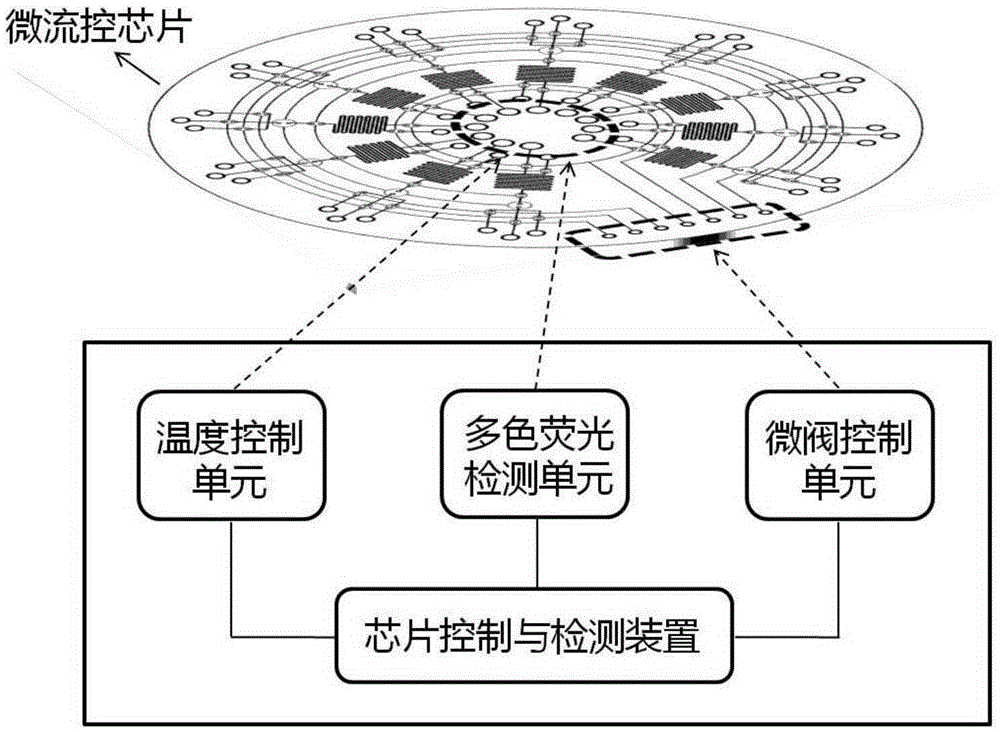
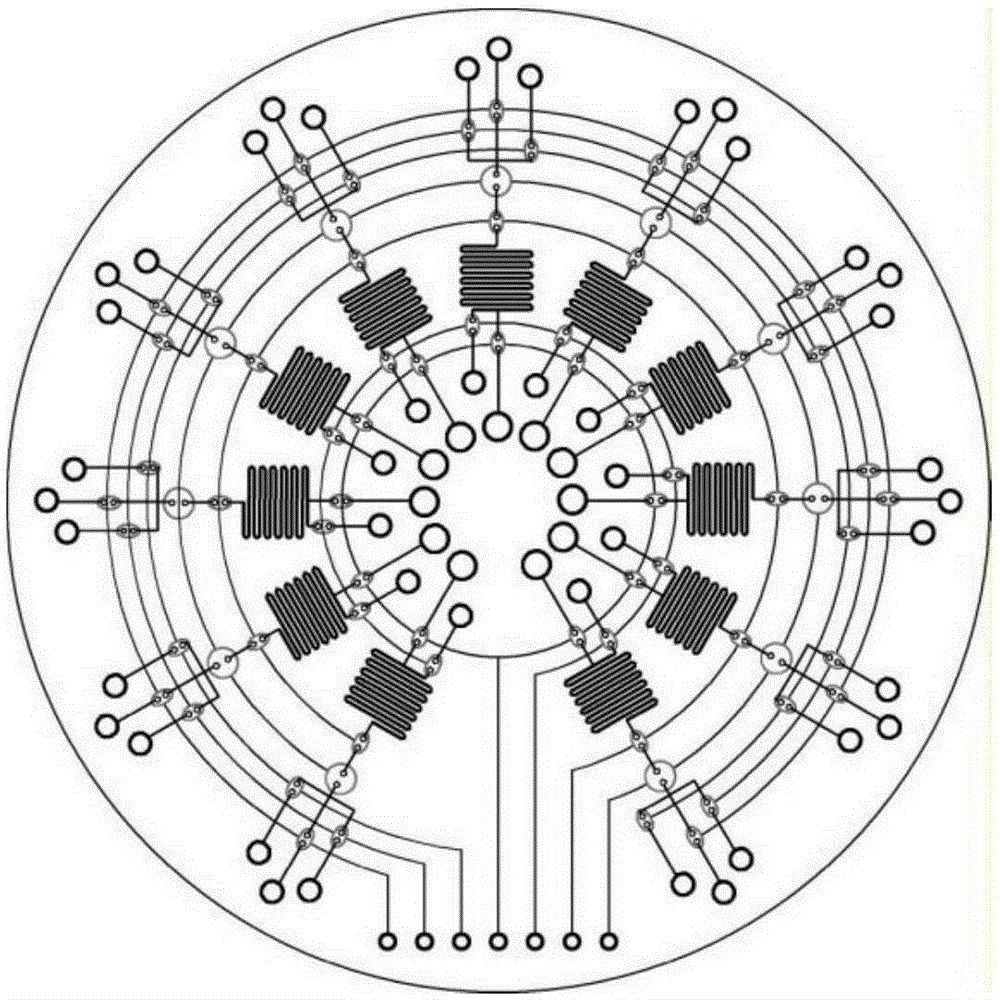
Microfluidic chip, detection system and device used for rapid DNA detection
InactiveCN105296349AImprove reliabilityGood repeatabilityBioreactor/fermenter combinationsBiological substance pretreatmentsFluid controlFluorescence
The present invention discloses a microfluidic chip for rapid DNA detection, and the chip includes a liquid storage unit, a nucleic acid extraction unit, an amplification and detection unit and a fluid control unit. The present invention further discloses a system and a device comprising the microfluidic chip for rapid DNA detection. The microfluidic chip can achieve repeated and automated nucleic acid extraction and fast and efficient PCR amplification and real-time fluorescence detection. At the same time, by use of a disc array structure, a plurality of samples can be analyzed in parallel, so that the method is expected to be used in scientific research and clinical body fluid sample DNA rapid detection.
Owner:融智生物科技(青岛)有限公司 +1
PCR diagnostics of dermatophytes and other pathogenic fungi
InactiveUS20100311041A1Sugar derivativesMicrobiological testing/measurementFungal microorganismsSkin specimen
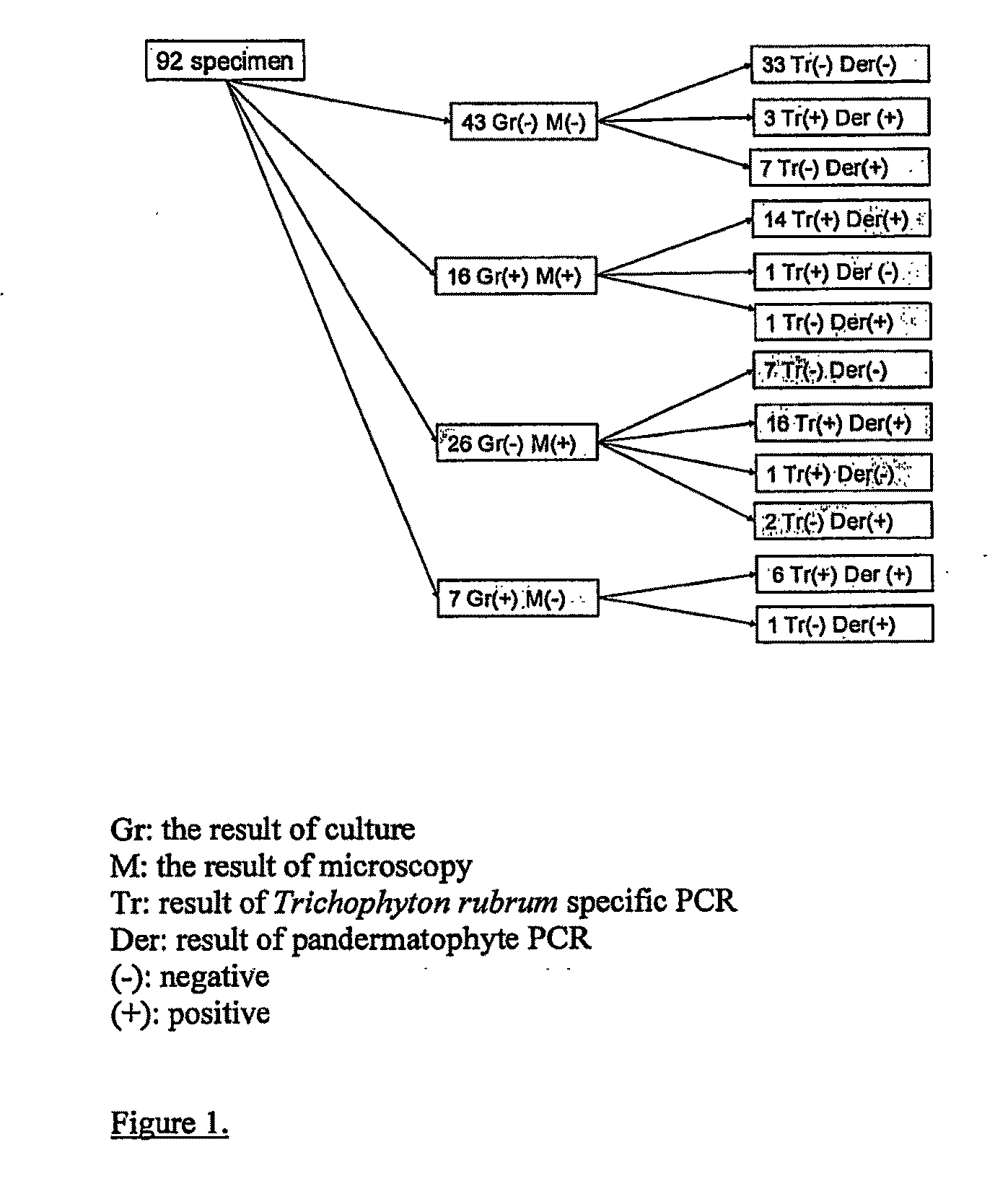
Dermatophytes which belong to one of the three genera Epidermophyton, Trichophyton and Microsporum are the main cause of fungal infections of skin, hair and nails. Traditional diagnostic procedures consist of microscopy and culture, but due to the slow growth rate of dermatophytes typically two to four weeks are needed before a final diagnosis is obtained. The present invention is a rapid DNA extraction method extracting nucleic acids from fungi (e.g. dermatophytes and other pathogenic fungi) which can be performed from directly on hair, nail or skin specimens from humans, from naturally or experimentally infected animals or from cultured fungal colonies for the use in PCR amplification and detection assays. The present invention also includes specific primer sets for detection of any dermatophyte and for species specific detection of Trichophyton rubrum and Epidermophyton floccosum by PCR and a kit for diagnosing fungal infections.
Owner:STATENS SERUM INST
Microfluidic systems with electronic wettability switches
InactiveUS20110266151A1Improve performanceEasy to processSludge treatmentVolume/mass flow measurementEngineeringSurface structure
The present invention concerns microfluidic systems with printed surface structured electronically controllable wettability switches for efficient manipulation of small amounts of fluids. The high performance microfluidic systems of the invention can be used in many applications, e.g. in rapid DNA separation and sizing, cell manipulation, cell sorting and molecule detection.
Owner:CELOXIO
Convenient and rapid DNA extracting method of peanut health tissues and diseased tissues
ActiveCN101805730AReduce dosageAssisted breedingSugar derivativesSugar derivatives preparationSingle sampleMaterials preparation
The invention relates to a convenient and rapid DNA extracting method of peanut health tissues and diseased tissues. The method comprises two steps of: sample materials preparation: testing the leaf and stem tissues, shell tissues or peanut cotyledons, the weight of which is 3-10 mg, adding the sample materials into a 1.5ml centrifugal tube preset with an alkaline lysate or enzymolysis liquid, and grinding by a grinding rod till the visible particles do not exist; DNA extracting liquid preparation: treating the ground sample materials by an alkaline decoction method or a enzymolysis method to obtain the DNA extracting liquid. The method is also suitable for peanut health tissues and diseased tissues. Compared with the DNA extracting method, the invention has short required time, the required time of single sample by the alkaline decoction method does not exceed 20min, and the required time of single sample by the enzymolysis method does not exceed 0.5h; the dosage of the sample is little, and each sample only needs 3-10mg; the method has no need of liquid nitrogen to grind, therefore the cost is low, and the requirements of peanut molecule marker auxiliary breeding and transgene breeding are satisfied. The invention has no influence on the plant growth or seed germination, and has significant meaning on the practicality of peanut molecule breeding technology.
Owner:SHANDONG PEANUT RES INST
Methods of amplifying and sequencing nucleic acids
An apparatus and method for performing rapid DNA sequencing, such as genomic sequencing, is provided herein. The method includes the steps of preparing a sample DNA for genomic sequencing, amplifying the prepared DNA in a representative manner, and performing multiple sequencing reaction on the amplified DNA with only one primer hybridization step.
Owner:454 LIFE SCIENCES CORP
Rapid DNA extraction kit of medicinal material
The invention discloses a rapid DNA extraction kit of a medicinal material, belongs to the field of the molecular biology, and relates to the traditional Chinese medicine and traditional Chinese medicinal material identification field. The kit comprises a solution A, a solution B, a reagent C and a user manual; and the solution A is composed of a solute triton-100 and a solvent water, the concentration of the solute is 1%, and the final concentration of the solute in the solution A 1%. The solution B is composed of 1M of a solute Tris-HCl and the solvent water, the concentration of the Tris-HCl in the solution B is 1M, the pH value of the solution B is 8.0, the concentration of the solute is 0.1M, and the final solution of the solute in the solution B is 0.1M. The reagent C is composed of NaOH and polyvinylpyrrolidone (PVP, M=40), the final concentration of NaOH in the solution A is 0.5M, and the final concentration of the PVP in the solution A is 1%. The kit is a medicinal material genome DNA extraction product developed for rapidly discriminating the medicinal material, and can be used for rapidly extracting plant and animal medicinal material genome DNA. The kit has the advantages of few operation steps, fast extraction speed, low price, no need of a large device, large-scale high-flux extraction and the like, and is very suitable for rapid detection and identification of the medicinal material DNA.
Owner:INST OF CHINESE MATERIA MEDICA CHINA ACAD OF CHINESE MEDICAL SCI
Eukaryotic cell nucleic acid extraction method
InactiveCN106350512AQuick extractionReduce pollutionMicrobiological testing/measurementDNA preparationCell membraneRapid dna
The invention discloses a eukaryotic cell nucleic acid extraction method and provides a nucleic acid extraction method which comprises the following steps of: uniformly mixing a to-be-tested sample with alkaline liquid, and performing reaction at a high temperature, thereby obtaining nucleic acid. Experiments prove that compared with an existing DNA extraction method, the method provided by the invention has the advantages that operation steps such as repeated cleaning, warm bath and centrifugation are removed; through creating an alkaline high-temperature liquid environment around the sample, membrane rupture and protein degeneration of eukaryotic cells are accelerated, the extraction efficiency of eukaryon genome DNA is greatly improved, and rapid DNA extraction is realized. The nucleic acid extraction method has no special requirements on aspects of sample types and laboratory conditions, is wide in application range and is suitable for popularization and application.
Owner:BGI GENOMICS CO LTD
Systems and Processes for Rapid Nucleic Acid Extraction, Purification, and Analysis
Disclosed are processes and kits for rapid nucleic acid extraction from a nucleic acid-containing material, such as a bone, tooth or semen sample. For bone and tooth process involves providing the nucleic acid-containing material in a form suitable for nucleic acid extraction, adding a lysis buffer to the nucleic acid-containing material to obtain a mixture, mixing the mixture in a manner equivalent for about 30 seconds or longer and separating the mixture by centrifugation to obtain a liquid supernatant. The liquid supernatant contains the extracted nucleic acids which can be used for analysis including STR profiling by conventional or rapid DNA analysis. For semen the processes and kits involve applying an appropriate amount of sperm disruptive agent.
Owner:ANDE CORP +1
STR typing method for loca D8S1179 based on next generation sequencing
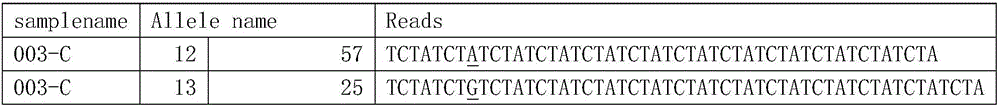
The invention relates to the field of molecular biology and particularly discloses a specific primer pair for amplifying a gene D8S1179. The specific primer pair comprises D8S1179-FP:5'-TTTGTATTTCATGTGTACATTCGTAATTC-3' and D8S1179-RP:5'-ACCTATCCTGTAGATTATTTTCACTGATTG-3'. The invention further provides an STR typing method for a loca D8S1179 based on next generation sequencing and particularly provides a primer combination for being connected with a proper adaptor. The STR typing method is applied to the STR typing of the loca D8S1179. According to the primer pair capable of amplifying the gene D8S1179, after concentrations of different sample products subjected to PCR amplification, the equivalent mixing is carried out, and a genome library is constructed according to a BIOO RAPID DNA kit; the library is input into a computer by virtue of a next generation sequencer, data is subjected to bioinformatics analysis, and the typing results such as times of repetition of NDA molecules with same length, SNP of the loca and the difference among side wings of the loca of D8S1179 can be found.
Owner:BEIJING CENT FOR PHYSICAL & CHEM ANALYSIS
Detection of extracellular tumor-associated nucleic acid in blood plasma or serum using nucleic acid amplification assays
InactiveUS20050176015A1Antibody mimetics/scaffoldsMicrobiological testing/measurementDiseaseNeoplasm
This invention relates to detection of specific extracellular nucleic acid in plasma or serum fractions of human or animal blood associated with neoplastic or proliferative disease. Specifically, the invention relates to detection of nucleic acid derived from mutant oncogenes or other tumor-associated DNA, and to those methods of detecting and monitoring extracellular mutant oncogenes or tumor-associated DNA found in the plasma or serum fraction of blood by using rapid DNA extraction followed by nucleic acid amplification with or without enrichment for mutant DNA. In particular, the invention relates to the detection, identification, or monitoring of the existence, progression or clinical status of benign, premalignant, or malignant neoplasms in humans or other animals that contain a mutation that is associated with the neoplasm through detection of the mutated nucleic acid of the neoplasm in plasma or serum fractions. The invention permits the detection of extracellular, tumor-associated nucleic acid in the serum or plasma of humans or other animals recognized as having a neoplastic or proliferative disease or in individuals without any prior history or diagnosis of neoplastic or proliferative disease. The invention provides the ability to detect extracellular nucleic acid derived from genetic sequences known to be associated with neoplasia, such as oncogenes, as well as genetic sequences previously unrecognized as being associated with neoplastic or proliferative disease. The invention thereby provides methods for early identification of colorectal, pancreatic, lung, breast, bladder, ovarian, lymphoma and all other malignancies carrying tumor-related mutations of DNA and methods for monitoring cancer and other neoplastic disorders in humans and other animals.
Owner:PENN STATE RES FOUND
Application of plant leaf grinding device for high-throughput rapid DNA extraction
InactiveCN105296469AImprove crushing effectEasy extractionBioreactor/fermenter combinationsBiological substance pretreatmentsThermal insulationEngineering
The invention relates to an application of a plant leaf grinding device for high-throughput rapid DNA extraction, and belongs to the technical field of biology. The grinding device comprises: a pedestal, a handle, and a plurality of grinding heads and connecting rods, wherein the pedestal is cuboid, is made of a thermal insulation material, and is a device for fixing the connecting rods and the handle; the upper part of the pedestal is connected to the handle; the lower part of the pedestal is connected to the connecting rods; and the lower parts of the connecting rods are connected to the grinding heads. The 96 grinding heads are connected to the base of the pedestal through the 96 connecting rods in an arrangement manner of 8 rows and 12 lines, wherein the transverse distance between two adjacent connecting rods is 0.37-0.9 cm and the longitudinal space distance between two adjacent connecting rods is 0.37-0.9 cm to match a 200 muL 96-hole PCR board and a 96-hole PCR device. For the application of the plant leaf grinding device for high-throughput rapid DNA extraction, whole sample crushing on the 96-hole PCR board can be completed in one step; a requirement of rapid DNA extraction and direct PCR experiment is met; and grinding time is greatly saved and labor requirement is lowered.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
Simple and fast extraction method for DNA in grape wine
InactiveCN101210032AHigh purityHigh yieldSugar derivativesMicrobiological testing/measurementThermal insulationGrape wine
The invention discloses a simple and rapid DNA extraction method in wine, which comprises the following steps: A. 5g Silica is added with 5ml aqueous suspension for Silica precipitation to be evenly mixed after being cleaned by pure water and is set at the temperature of 4 DEG C for standby; B. wine with best adsorption conditions is then produced; C. the DNA of the wine is adsorbed and recycled: (1) Silica suspension is added to the wine mixture; (2) conservation is done for 1-2 hours under the temperature of 4DEG C, with constant shaking and oscillation; (3) centrifugation is done for 5 min under the temperature of 4DEG C with the amount of 5000g; (4) supernatant is removed and the Silica deposit is washed for 1-2 times by using washing fluid containing 0.1M Tris-HCl, pH 8.0, 1 percent of CTAB and 1M NaCl; (5) the centrifugation is done for 5 min with the amount of 5000g to collect the Silica deposit; (6) the Silica deposit is added with the pure water with the equivalent amount as the deposit and then well shaked and done with DNA heat elution after 5-10 min thermal insulation at the temperature of 65 DEG C; (7) centrifugation (10000g, 5min) is done to collect the supernatant containing DNA; (8) the supernatant in step (7) is extracted by chloroform with the same volume and the centrifugation (10000g, 3min) is done to collect top phase containing DNA; (9) the DNA in the supernatant is recycled and concentrated by using isopropanol precipitation; (10) the recycled wine DNA is then dissolved in 50 Mu lota TE (10mM Tris-HCl, pH 8.0) buffering agent.
Owner:HENAN AGRICULTURAL UNIVERSITY
Novel rapid DNA separation and sequencing method for mitochondrial genome and kit
The invention discloses a novel rapid DNA separation and sequencing method for a mitochondrial genome and a kit. A linear chromosome sequence is degraded by Exonuclease III or Plasmid-Safe ATP-Dependent DNase, and then mitochondrial genome DNA is enriched for high-throughput sequencing. The mitochondrial DNA sequencing method is applied to detection of the mitochondrial genome and particularly detection of the mitochondrial genome without a consensus sequence.
Owner:ZHEJIANG UNIV
Rapid DNA extraction method of citrus leaves, and application thereof in detection of citrus yellow shoot diseases
ActiveCN110184266AEasy to operateReduce processing timeMicrobiological testing/measurementDNA preparationDiseaseWater baths
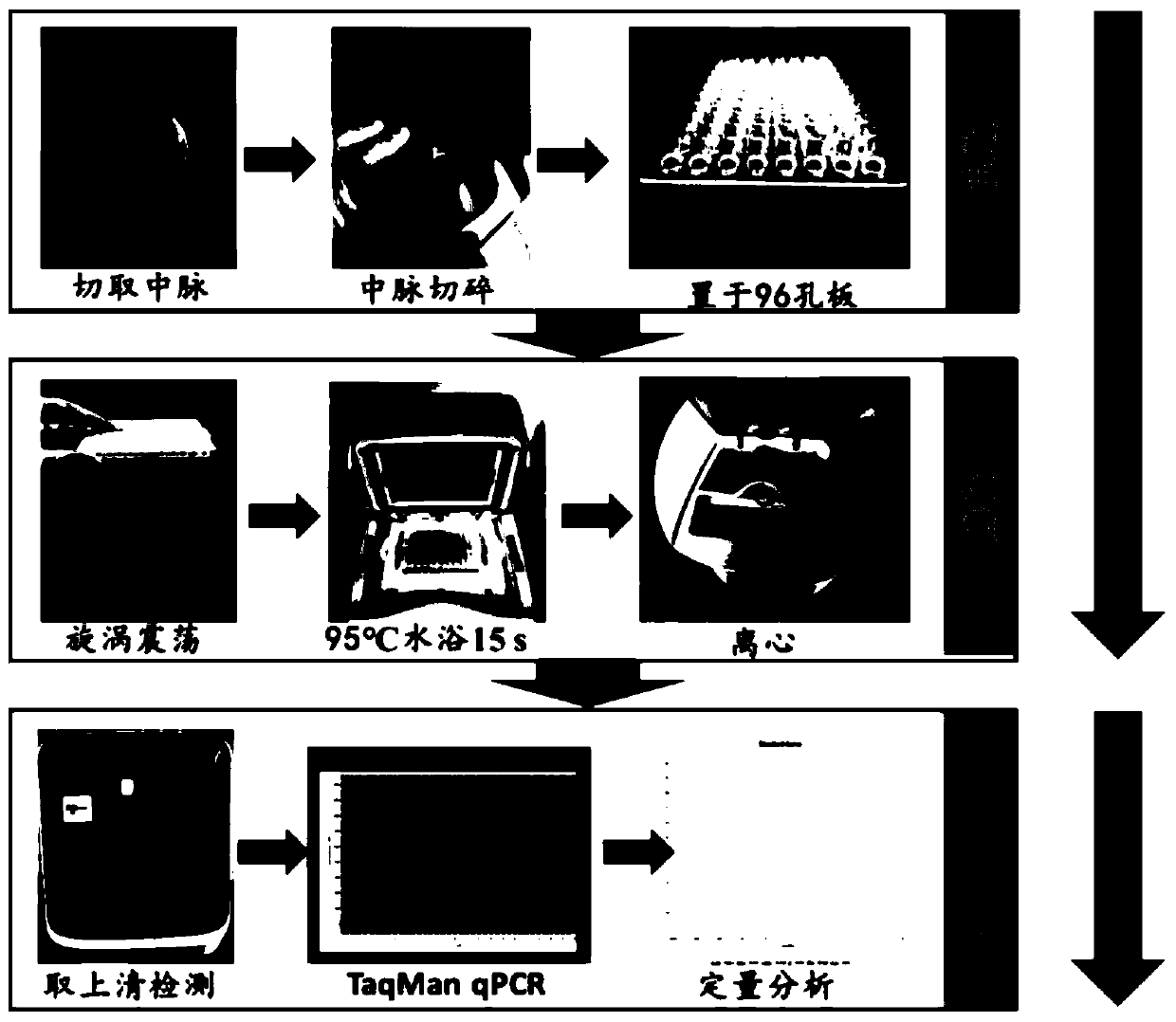
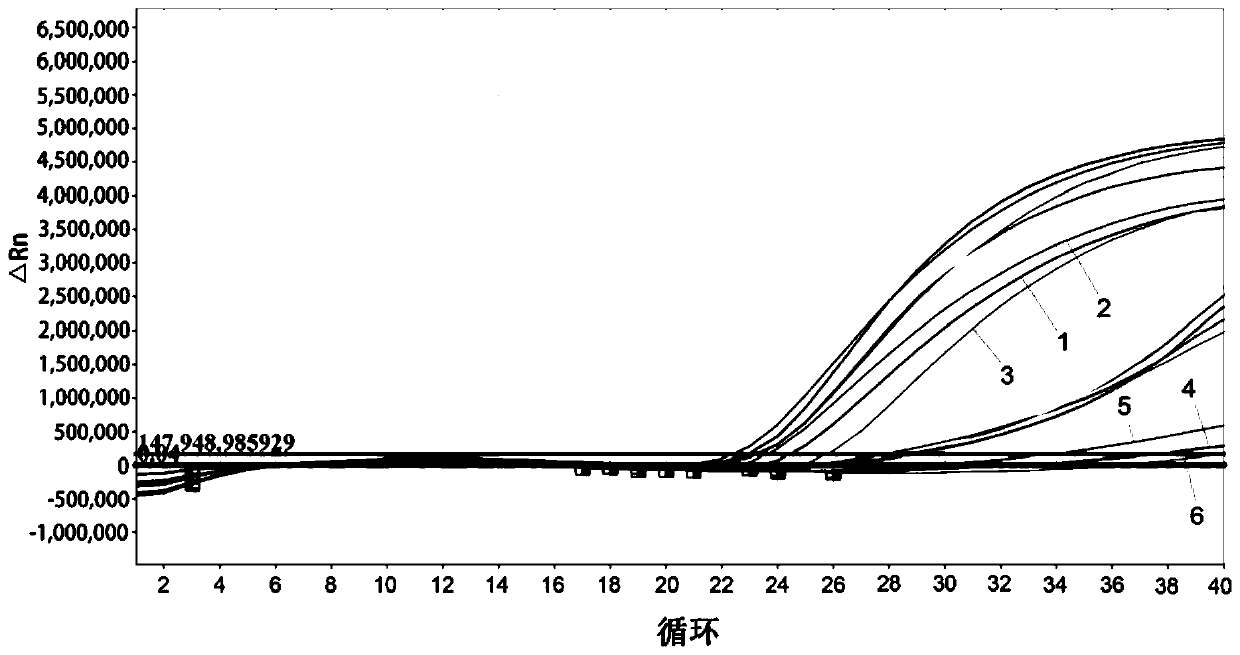
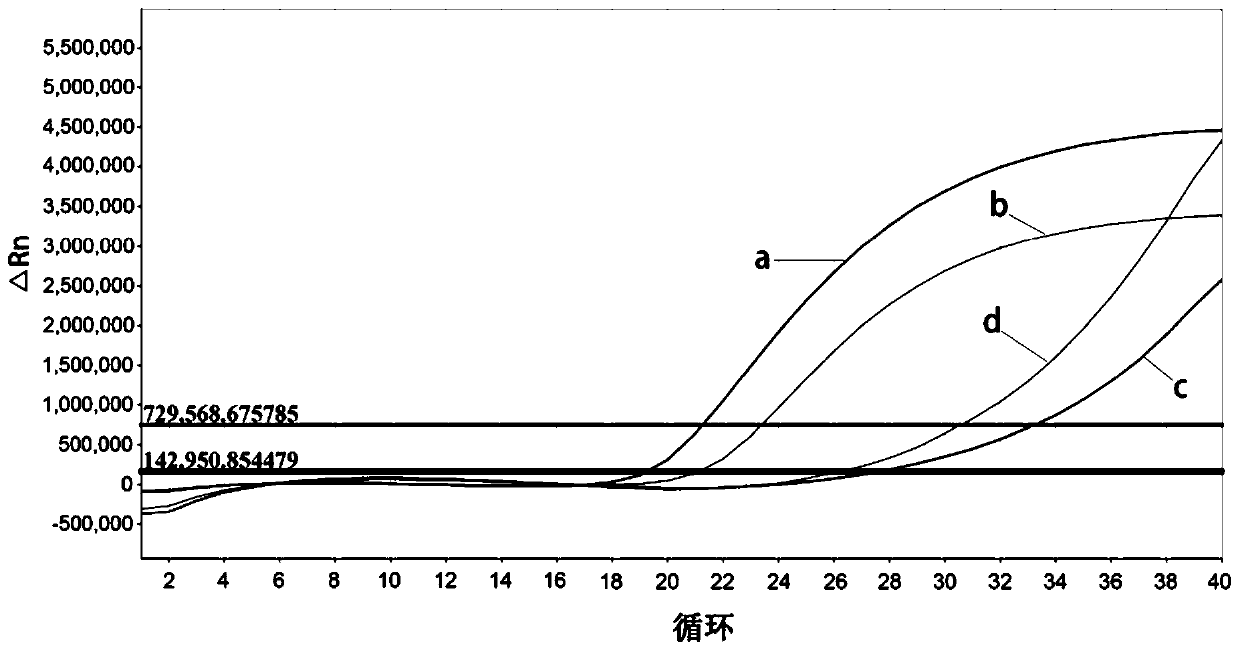
The present invention relates to detection of citrus diseases and particularly relates to a rapid DNA extraction method of citrus leaves, and an application thereof in a high-throughput molecular detection of citrus yellow shoot diseases. The rapid DNA extraction method comprises the following steps: petioles and leaf midribs of the citrus leaves from citrus plants to be tested are cut and crushed, 40 [mu]L of lysate is added into each 30 mg of a sample, vortexing vibrating is conducted for 10-15 s, water bath at 95 DEG C is conducted for 15 s, then 0.1 mol / L of Tris-HCL with a pH of 8 and a volume 4 times of the lysate is added, the vortexing vibrating is conducted for 10-15 s, and centrifuging is conducted to obtain a supernatant, a DNA solution; and a formula of the lysate is as follows: 0.5 mol / L NaOH and 1% TritonX-100. The method is combined with qPCR to achieve the high-throughput rapid molecular detection of the citrus yellow shoot diseases, and provides a favorable tool for large-scale and accurate diagnosis of the citrus yellow shoot diseases.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
Detection kit for budesonide metabolism marker and detection method and application thereof
PendingCN113667743ASimplify the sequencing workflowSimplify timeMicrobiological testing/measurementDNA/RNA fragmentationRapid dnaBudesonide
The invention discloses a detection kit for a budesonide metabolism marker and a detection method and application thereof. The kit designs a specific amplification primer and a sequencing primer aiming at the polymorphism of GLCCI1A-1106G, and the kit comprises the following components: a sample treating fluid, magnetic beads, an amplification reagent 1, an amplification reagent 2, a GLCCI1A-1106G sequencing primer and a positive control. The detection kit is mainly optimized from two aspects: on one hand, a mode of extracting and amplifying in the same tube is adopted, so that the risk of nucleic acid loss due to repeated tube transfer during extraction is avoided; on the other hand, rapid isothermal amplification is carried out by adding an anti-inhibitor; the invention aims to detect the gene polymorphism of the curative effect of budesonide by combining rapid DNA (deoxyribonucleic acid) preparation, constant-temperature PCR (polymerase chain reaction) amplification and pyrosequencing technologies, and provides suggestions from the gene perspective for clinical personalized medication.
Owner:菲思特(上海)生物科技有限公司
Rapid DNA (deoxyribonucleic acid) extraction kit with magnetic beads for fluorescent quantitative PCR (polymerase chain reaction) detection and extraction method of rapid DNA extraction kit
PendingCN114657231AReasonable ratioFully lysedMicrobiological testing/measurementMicroorganism based processesLysisMagnetic bead
The invention discloses a rapid DNA (deoxyribonucleic acid) extraction kit with magnetic beads for fluorescent quantitative PCR (polymerase chain reaction) detection and an extraction method thereof, and belongs to the technical field of bioengineering. The rapid DNA extraction kit comprises a rapid DNA extraction kit body; the rapid DNA extraction kit comprises a lysis binding solution LB, a washing solution W1, a washing solution W2, a nucleic acid elution reaction solution EB, magnetic beads MB and protease K. The extraction method comprises a manual nucleic acid extraction method or a method for realizing high-flux automatic DNA extraction by matching with an automatic instrument. The rapid DNA extraction reagent with the magnetic bead amplification function can meet DNA extraction of various sample types such as whole blood, plasma and swabs, an extracted product can be directly applied to fluorescent quantitative PCR amplification detection reaction, separation of magnetic beads and nucleic acid is not needed, downstream detection is not affected, and nucleic acid extraction or purification can be achieved more simply, more sensitively and more rapidly.
Owner:珠海宝锐生物科技有限公司
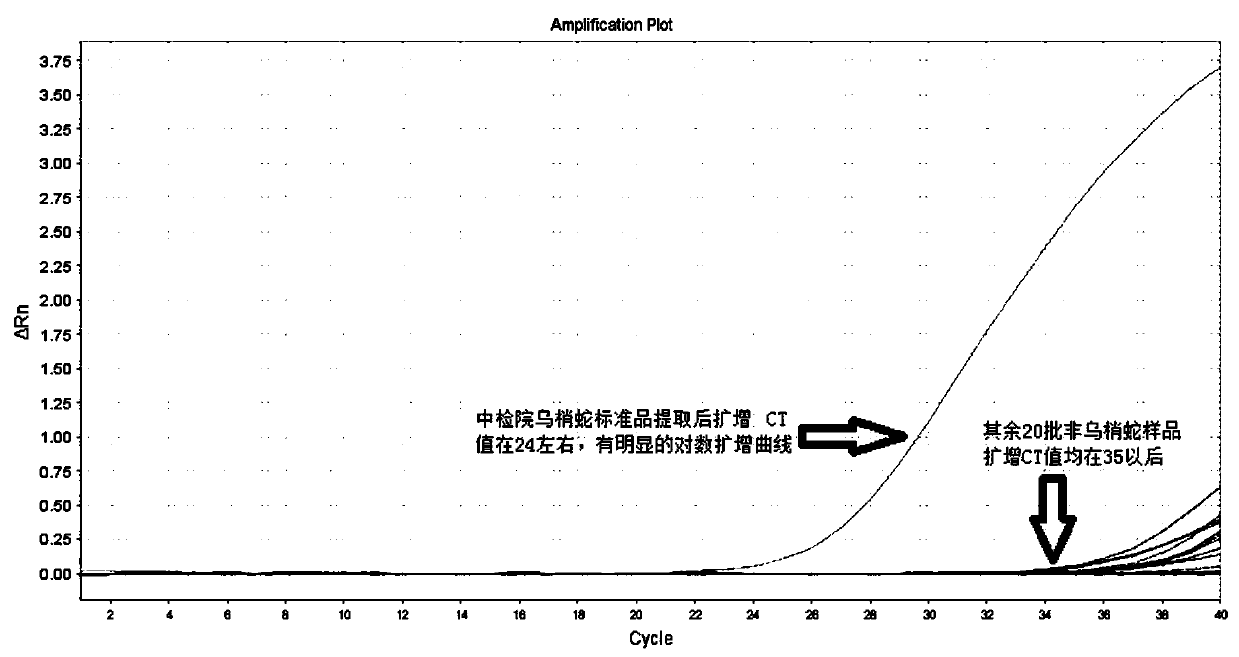
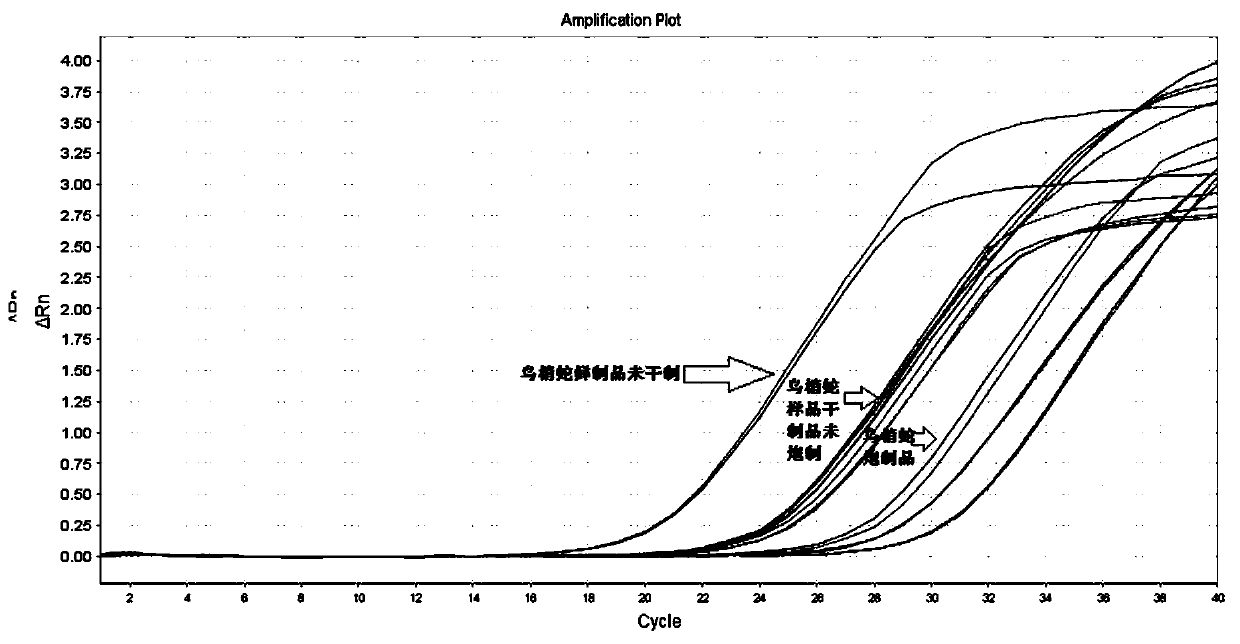
Real-time fluorescent PCR method for identifying authenticity of garter snake traditional Chinese medicinal material by adopting specific primers
PendingCN109943650AStrong specificityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceElectrophoresis
The invention belongs to the technical field of identification of traditional Chinese medicines and traditional Chinese medicinal materials, and provides a real-time fluorescent PCR method for identifying the authenticity of a garter snake traditional Chinese medicinal material by adopting specific primers, aiming at solving the problems that the existing garter snake detection method is easy to have a false positive result and cannot be applied to market rapid detection and the like. A specific primer sequence used for identifying the authenticity of the garter snake traditional Chinese medicinal material is shown as SEQ ID NO: 1 and SEQ ID NO: 2; a probe sequence is shown as SEQ ID NO: 3. The identification of the authenticity of samples can be finished through rapid DNA extraction and real-time fluorescent PCR specific amplification. The real-time fluorescent PCR method for identifying the authenticity of the garter snake traditional Chinese medicinal material by adopting the specific primers has the advantages that the primer probe double specificity has higher specificity; the fluorescence detection sensitivity is higher; the detection time of the method is short without an electrophoresis process; a pharmacopoeia detection method needs about 4 hours, and the real-time fluorescent PCR method for identifying the authenticity of the garter snake traditional Chinese medicinalmaterial by adopting the specific primers only needs 1.5 hours; the required instrument and equipment are less, and the method can be applied to a rapid detection vehicle service market, and the like.
Owner:太原市食品药品检验所
Gene polymorphism detection kit for second-generation antipsychotic drug metabolism marker as well as detection method and applicationof gene polymorphism detection kit
PendingCN113755577AAvoid the risk of lossSimplify timeMicrobiological testing/measurementDNA/RNA fragmentationMedicinePharmaceutical drug
The invention discloses a gene polymorphism detection kit for a second-generation antipsychotic drug metabolism marker as well as a detection method and application of the gene polymorphism detection kit. The detection kit is characterized in that a specific amplification primer and a sequencing primer are designed aiming at the polymorphism of ANKKI 2317G > A, the kit comprises the following components: a sample treating fluid, magnetic beads, an amplification reagent 1, an amplification reagent 2, an ANKKI (2317G > A) sequencing primer and a positive control. The gene polymorphism detection kit is mainly optimized from two aspects: on one hand, a mode of extracting and amplifying in a same tube is adopted, so that the risk of nucleic acid loss due to repeated tube transfer during extraction is avoided; on the other hand, rapid isothermal amplification is carried out by adding an anti-inhibitor; the gene polymorphism detection kit aims to detect the gene polymorphism of the second-generation antipsychotic drug by combining rapid DNA preparation, constant-temperature PCR amplification and pyrosequencing technologies, and provides suggestions from the gene perspective for clinical personalized medication.
Owner:菲思特(上海)生物科技有限公司
A simple and rapid method for DNA extraction from peanut healthy tissue and diseased tissue
ActiveCN101805730BReduce dosageAssisted breedingSugar derivativesSugar derivatives preparationDiseaseTwo step
The invention relates to a simple and quick method for extracting DNA from peanut healthy tissues and diseased tissues, which includes two steps. The preparation of sample materials is to take leaves, stem tissues, shell tissues or peanut cotyledons with a weight of 3-10 mg for the test; the sample materials are added to the preset In a 1.5ml centrifuge tube with alkali lysate or enzymolysis solution, grind it with a grinding rod until there are no visible particles; the preparation of DNA extraction refers to the use of alkali boiling method or enzymolysis method to process the above ground sample material, Obtain sample DNA extract. This law is equally applicable to peanut healthy organizations and peanut diseased organizations. Compared with the existing DNA extraction methods, the present invention consumes less time. The time for a single sample in the alkali boiling method does not exceed 20 minutes, and the time for a single sample in the enzymatic hydrolysis method does not exceed 0.5 h; the amount of samples is small, and each sample only needs 3-10 mg; Nitrogen grinding, low cost, can meet the requirements of peanut molecular marker assisted breeding and transgenic breeding. It has no effect on plant growth or seed germination, and is of great significance for the practical application of peanut molecular breeding technology.
Owner:SHANDONG PEANUT RES INST
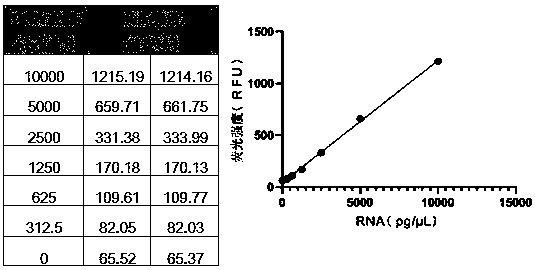
Universal method for rapidly quantifying RNA residue in DNA product
ActiveCN110389230AQualitatively accurateHigh sensitivityBiological testingFluorescence/phosphorescenceFluorescenceA-DNA
The invention discloses a universal method for rapidly quantifying RNA residues in a DNA product. According to the method, RNA is quantitatively detected by combining a specific nuclease with a RNA specific fluorescent dye. The method specifically comprises the following steps: digesting DNA by specific nuclease to reserve RNA, eliminating interference, and then quantitatively detecting RNA residues by using the RNA specific fluorescent dye; or, after the RNA is digested by the specific nuclease, quantitatively detecting the RNA by using the RNA specific fluorescent dye, and calculating a difference value before and after digestion to quantitatively detect the RNA residue. Compared with conventional AGE qualitative detection, the method provided by the invention is quantitative detection,and because the RNA specific fluorescent dye has high sensitivity to RNA and is theoretically less influenced by the length difference of RNA fragments, the method provided by the invention has the advantages of good quantitative linearity, high sensitivity, small RNA fragmentation interference and good accuracy; compared with a conventional RT-qPCR mode to indirectly quantifies the RNA residues,the method directly quantifies the RNA residues, is simple and rapid in experimental process and low in cost; theoretically, the method directly quantifies the RNA with more accuracy, and has low requirements on experimenters.
Owner:无锡生基医药科技有限公司
Method for detecting various microorganisms by integrated multiple nest type PCR one-step method
PendingCN110273014AAvoid individual locus differencesUniform GC contentMicrobiological testing/measurementDNA/RNA fragmentationGenomic DNAGel electrophoresis
The invention discloses a method for detecting various microorganisms by an integrated multiple nest type PCR one-step method. The method comprises the steps of performing differentiation designing of a universal primer and a specific primer of NM-PCR based on the characteristics of DNA bar code genes of microorganisms to be detected, establishing an NM-PCR one-step detection system, namely performing parameter optimization by using the genomic DNA of the microorganisms to be detected as a PCR reaction formwork, performing quick DNA extraction on microorganism genomes in samples to be detected, performing a NM-PCR amplification reaction by using the genomic DNA in the samples as a formwork, and performing gel electrophoresis detection on PCR reaction products. The method disclosed by the invention concurrently has specificity of the nest type PCR and high flux of multiplex PCR, two-time PCR processes are completed only through one-step PCR reaction, traditional PCR multi-step operations are avoided, the microbial pollution situations of clinical samples such as environment water sources and foods can be quickly and massively detected, the specificity is high, the flus is high, the detection time can be greatly shortened, and the cost can be greatly reduced.
Owner:SHANGHAI JIAO TONG UNIV
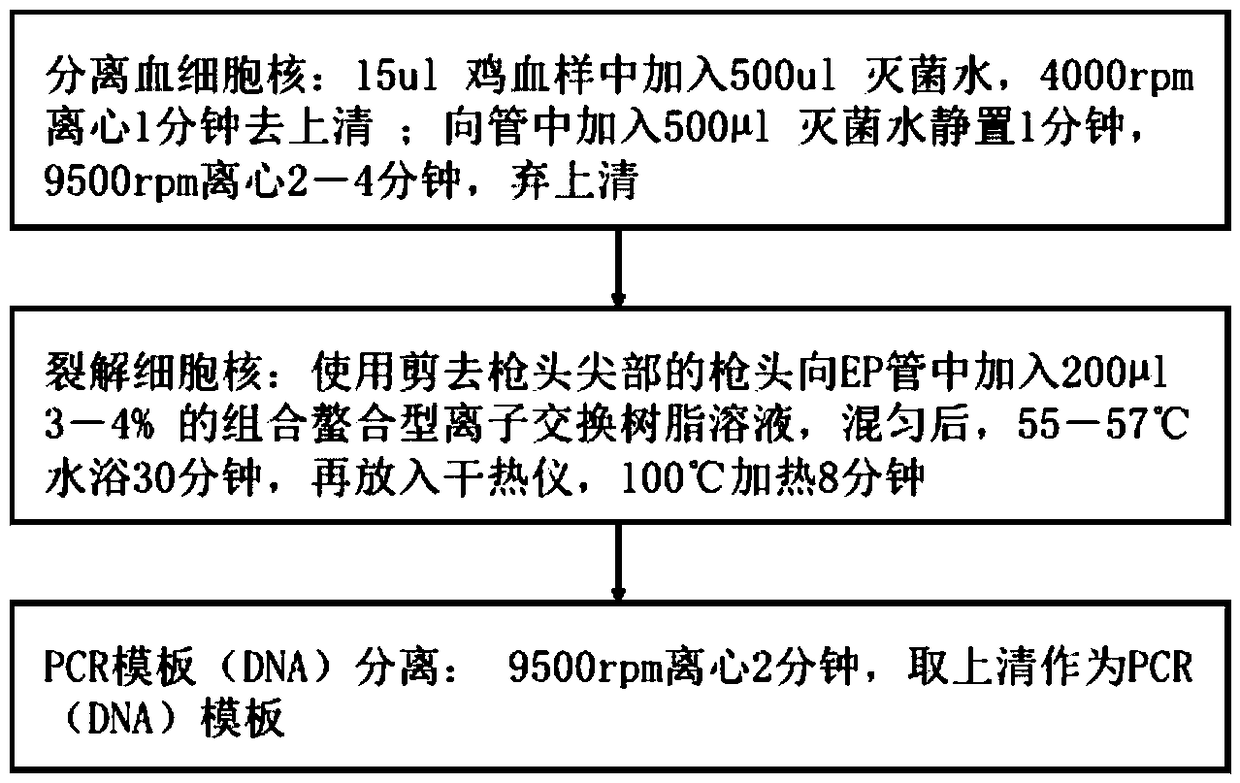
Rapid and low-cost method for separating PCR template from chicken blood
InactiveCN109402112AEasy to separateLow costMicrobiological testing/measurementDNA preparationWater bathsCentrifugation
The invention discloses a rapid and low-cost method for separating a PCR template from chicken blood. The rapid and low-cost method for separating the PCR template from the chicken blood comprises thesteps that A, blood corpuscle nucleuses are separated from a blood sample, specifically, a fresh and frozen anticoagulant chicken blood sample is added into a 1.5 mL EP pipe, and 4000rpm centrifugation is carried out for one minute and supernatant is removed; and 500 [mu]l sterile water is added into the pipe to stand for one minute, 9500rpm centrifugation is carried out for three minutes, and the supernatant is abandoned; B, the corpuscle nucleuse dissociation is carried out, specifically, a 3-4% chelated type ion exchange resin solution shaken up by the 200 [mu]l sterile water is added intosediment by using a spearhead with the cut tip part, after mixing, water bath at 55 DEG C-57 DEG C is carried out for 30 minutes, then, a dry heat meter is placed in, and heating is carried out at 100 DEG C for 8 minutes; and C, PCR template separation is carried out, specifically, 9500rpm centrifugation is carried out for two minutes, and the supernatant is taken as a PCR template. Compared witha template obtained by using an existing traditional DNA extraction method, the PCR template obtained by using the rapid and low-cost method for separating the PCR template from the chicken blood hassame effects for PCR technology detecting, the operation is simpler and easier to operate, the time-consuming is short (within one hour), the cost is low (about 0.31 yuan / sample), and the PCR template is suitable for various rapid DNA molecular detection experiments carried out by using PCR technology.
Owner:HUAZHONG AGRI UNIV
Rapid LAMP detection method for watermelon fusarium oxysporum
InactiveCN113186333AMeet testing needsGood repeatabilityMicrobiological testing/measurementMicroorganism based processesBiotechnologySpore
The invention discloses a rapid LAMP detection method for watermelon fusarium oxysporum. According to the method, a set of sensitive and specific LAMP primers are designed by taking a specific DNA fragment of fusarium oxysporum watermelon specialized type as a target, and a rapid LAMP detection system of the watermelon fusarium oxysporum is established by combining a rapid DNA extraction method. The lower detection limit of the LAMP system on FON-1 genome DNA is 10 pg / [mu]L, and the lower detection limit of the LAMP system on the plant bacterium carrying amount is 5 * 10<2> spores / g. According to the system, only about 90 minutes are needed from DNA extraction to detection result acquisition, and pathogenic bacteria hyphae and plant tissues can be used as detection materials. The reaction result can be distinguished by naked eyes. The method has the characteristics of simplicity in operation, high specificity, high sensitivity, high stability and wide applicability, and can provide technical support for rapid diagnosis of field watermelon fusarium oxysporum.
Owner:HUNAN AGRI BIOTECH RES CENT
Nested PCR (polymerase chain reaction) detection primer and detection method for Uromyces vignae
ActiveCN110951914AAchieving Specific DetectionStrong specificityMicrobiological testing/measurementAgainst vector-borne diseasesElectrophoresesAgarose electrophoresis
The invention relates to a nested PCR (polymerase chain reaction) detection primer and a detection method for Uromyces vignae. The nested PCR detection primer for the Uromyces vignae adopts a specificdetection primer for the Uromyces vignae to amplify genome DNA (deoxyribonucleic acid), and sequences an amplification product to obtain sequence information of a UV-ITS (ultraviolet-internal transcribed spacer) amplification product; the sequence information is used as a template to design the nested PCR primer, and the nested PCR primer is named as UV-MX to obtain the nested PCR detection primer for the Uromyces vignae as shown in SEQ ID NO: 1 and SEQ ID NO: 2. The nested PCR detection primer and the detection method for the Uromyces vignae are simple, convenient and rapid to operate; a result can be judged after the genome DNA of a sample to be detected is subjected to extraction, PCR amplification and conventional agarose electrophoresis; a rapid DNA extraction method is adopted in the whole detection process, and pathogenic bacteria do not need to be subjected to isolated culture, so that the detection time is greatly shortened; and the whole detection process can be generally finished within 6 hours.
Owner:HEILONGJIANG BAYI AGRICULTURAL UNIVERSITY
Gene polymorphism detection kit for predicting adverse reaction and curative effect of fluorouracil as well as detection method and application of gene polymorphism detection kit
PendingCN113755588ASimplify timeMicrobiological testing/measurementDNA/RNA fragmentationUracil in DNAFluorouracilum
The invention discloses a gene polymorphism detection kit for predicting adverse reaction and curative effect of fluorouracil as well as a detection method and application of the gene polymorphism detection kit. The detection kit is used for detecting gene polymorphism of fluorouracil metabolism related genes TYMS, GSTP1, NQO1 and MTHFR, the kit designs specific amplification primers and sequencing primers for the TYMS, GSTP1, NQO1 and MTHFR. The kit comprises the following components: a sample treating fluid, magnetic beads, an amplification reagent 1, an amplification reagent 2, the TYMS, the GSTP1, the NQO1, an MTHFR sequencing primer and a positive control. According to the gene polymorphism detection kit for predicting the adverse reaction and the curative effect of the fluorouracil as well as the detection method and application of the gene polymorphism detection kit, rapid DNA preparation, constant-temperature PCR amplification and pyrosequencing technologies are combined to detect the gene polymorphism for predicting the adverse reaction and curative effect of the fluorouracil, and suggestions from the gene perspective are provided for clinical personalized medication.
Owner:菲思特(上海)生物科技有限公司
A next-generation sequencing-based method for STR typing of the locus D8S1179
The invention relates to the field of molecular biology and particularly discloses a specific primer pair for amplifying a gene D8S1179. The specific primer pair comprises D8S1179-FP:5'-TTTGTATTTCATGTGTACATTCGTAATTC-3' and D8S1179-RP:5'-ACCTATCCTGTAGATTATTTTCACTGATTG-3'. The invention further provides an STR typing method for a loca D8S1179 based on next generation sequencing and particularly provides a primer combination for being connected with a proper adaptor. The STR typing method is applied to the STR typing of the loca D8S1179. According to the primer pair capable of amplifying the gene D8S1179, after concentrations of different sample products subjected to PCR amplification, the equivalent mixing is carried out, and a genome library is constructed according to a BIOO RAPID DNA kit; the library is input into a computer by virtue of a next generation sequencer, data is subjected to bioinformatics analysis, and the typing results such as times of repetition of NDA molecules with same length, SNP of the loca and the difference among side wings of the loca of D8S1179 can be found.
Owner:BEIJING CENT FOR PHYSICAL & CHEM ANALYSIS
Dual-multienzyme-mediation-based nucleic acid amplification detection method and reagent
PendingCN112501262AHigh detection sensitivityShort detection timeMicrobiological testing/measurementRapid identificationSingle strand
The invention relates to a dual-multienzyme-mediation-based nucleic acid amplification detection method. The method comprises the following steps: pre-amplification, freeze-dried redissolution, amplified detection and result analyzing. According to the method, a normal-temperature nucleic acid amplification technology is combined with a nest type amplification principle, a plurality of enzymatic reactions such as single-stranded DNA binding protein, DNA polymerase and DNA double-strand-opened enzyme are involved, DNA can be amplified by several million folds or more in 10 to 20 minutes under the constant-temperature condition of 30 DEG C to 45 DEG C, and rapid identification on the DNA can be achieved by being matched with a fluorescence detection technique. The method disclosed by the invention is simple in operation, short in time and low in instrument requirements and is applicable to rapid DNA diagnosis.
Owner:苏州晶睿生物科技有限公司
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com