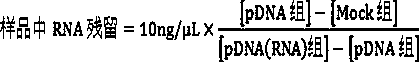Universal method for rapidly quantifying RNA residue in DNA product
A product and fast technology, which is applied in measuring devices, material inspection products, and material analysis through optical means, etc., can solve problems such as complex experimental procedures, reducing the sensitivity and accuracy of AGE method, and interfering RNA band detection, etc., to achieve The experimental process is simple, the effect of fragmentation interference is small, and the cost is low
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
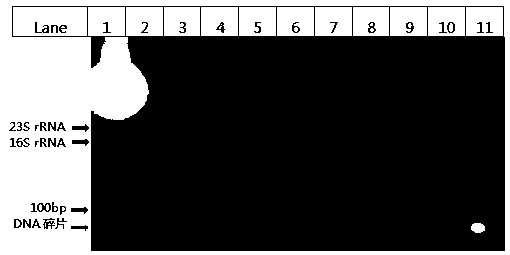
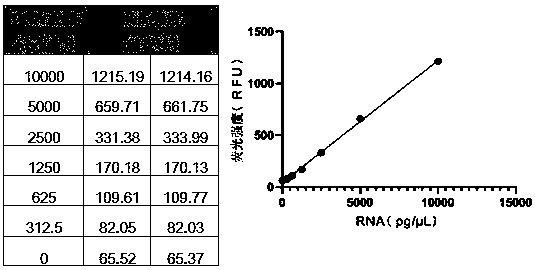
Image
Examples
Embodiment 1
[0033] 1. Preferred specific nuclease
[0034] The interference of DNase I (Thermo Scientific, EN0523) combined with RNase Inhibitor (Promega, N2611) and RNase A (Thermo Scientific, EN0531) alone on the fluorescence signal of RNA-specific nucleic acid dye Qubit RNA HS (Invitrogen, Q32852) was compared.
[0035] Prepare the following solutions as needed:
[0036] 1) DNase I Mix: 3.2µL DNase I, 16µL 10X Reaction Buffer with MgCl 2 , 0.8µL RNase Inhibitors, 60µL H 2 O (Nuclease-Free)
[0037] 2) RNase A Mix: 490TE Buffer+10µL RNase A
[0038] 3) TE Buffer (RI): 99µLTEBuffer, 1µLRNase Inhibitor
[0039] 4) QubitWorking Solution: 3µL QubitRNAHSReagent, 597µL QubitRNAHSBuffer
[0040] Prepare the experimental group:
[0041] 1) Blank group: 20µLTE buffer
[0042] 2) DNase Mix group: 10 µLTE buffer (RI), 10 µLDNase I Mix (DNase I final concentration 1U / µL)
[0043] 3) RNase Mix group: 10 µLTE buffer, 10 µL RNase AMix (the final concentration of RNase is 0.1 µg / µL)
[0044] Dig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com