Rapid Qualitative and Quantitative Determination of Saccharomyces cerevisiae in Additive Premix Samples
A technology for the qualitative determination of Saccharomyces cerevisiae, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, and material stimulation analysis. The effect of short cycle, good accuracy and high detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1: Rapid qualitative detection of Saccharomyces cerevisiae in premix samples:
[0056] This embodiment uses the gel recovery product of the purpose amplified fragment as a positive control; Sterile water is used as a negative control; In addition, two parts of feed samples 1# and 2#, Enterococcus faecalis, and Pichia pastoris collected from the market are used as samples to carry out .
[0057] A. Preparation of Template DNA
[0058] The template DNA of the feed sample was prepared by the glass bead method, and the specific steps were as follows:
[0059] (1) Take 1 g feed sample containing Saccharomyces cerevisiae in a 1.5 mL centrifuge tube, add 500 μL PBS to suspend the sample, and centrifuge at 2500 × g for 3 min;
[0060] (2) Discard the supernatant, add an appropriate amount of acid-washed glass beads, and shake vigorously for 2 minutes;
[0061] (3) Add 400 μL TE and an equal volume of Tris-saturated phenol, invert and mix well, and centrifuge at 15,00...
Embodiment 2
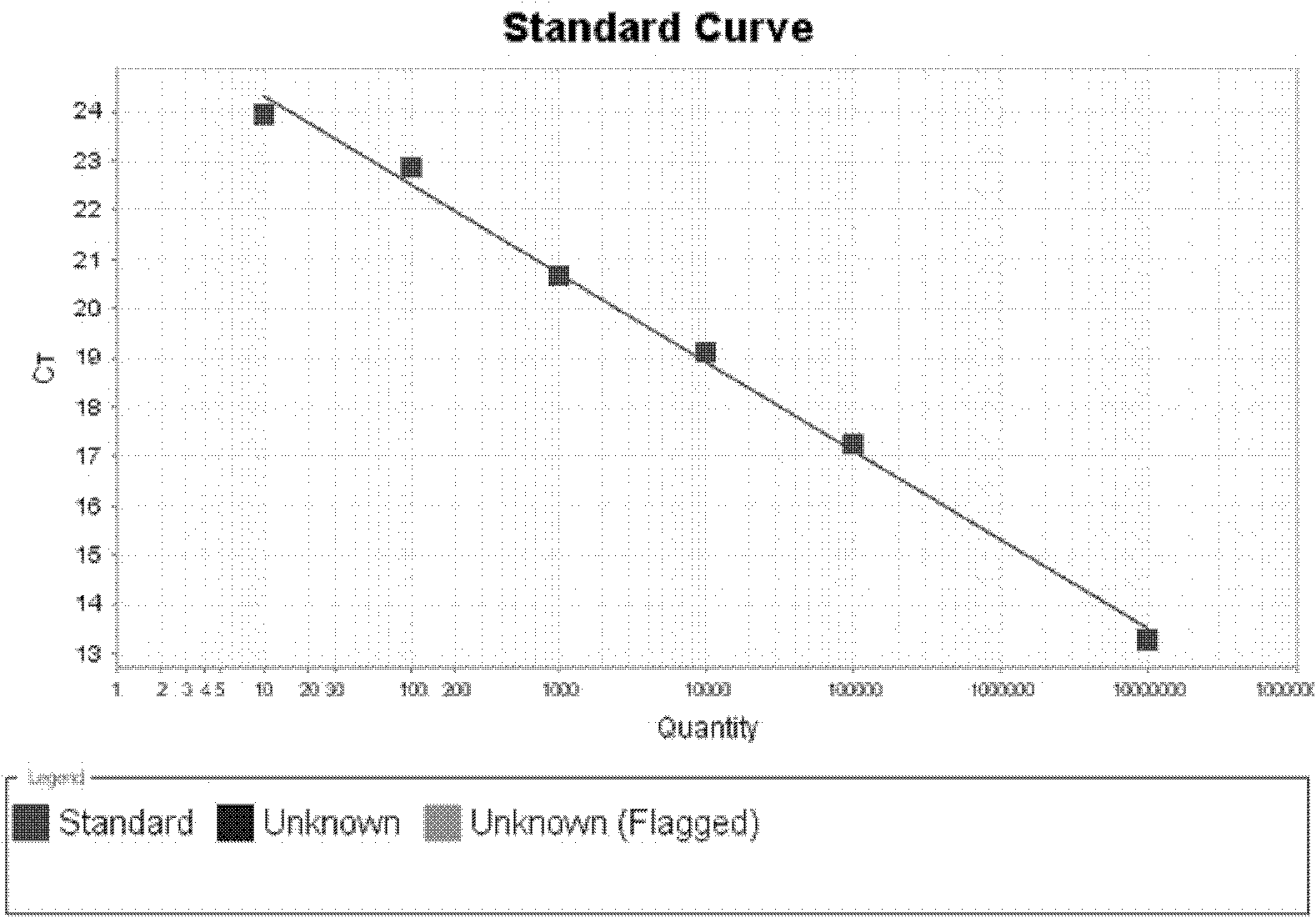
[0075] Example 2: Quantitative determination of Saccharomyces cerevisiae in premix samples
[0076] Quantitative determination of Saccharomyces cerevisiae was carried out on the different samples mentioned in Example 1. Its determination steps are as follows:
[0077] A. Preparation of total sample DNA
[0078] Use enzymatic method to break the yeast cell wall and extract the total DNA of the sample with the high-purity total DNA rapid extraction kit (spin column type): add 0.5-1.0 g of the sample to a 1.5 mL centrifuge tube, add 600 μL of sorbitol buffer, add 50 U of Lyticase, and mix well Treat at 30°C for 30 minutes, centrifuge at 1500×g for 10 minutes, discard the supernatant, collect the precipitate, extract the sample DNA according to the instructions of the high-purity total DNA rapid extraction kit, measure the concentration with a UV spectrophotometer, and detect it by 1% agarose gel electrophoresis The integrity of the DNA was then stored at -20°C.
[0079] B. Flu...
Embodiment 3
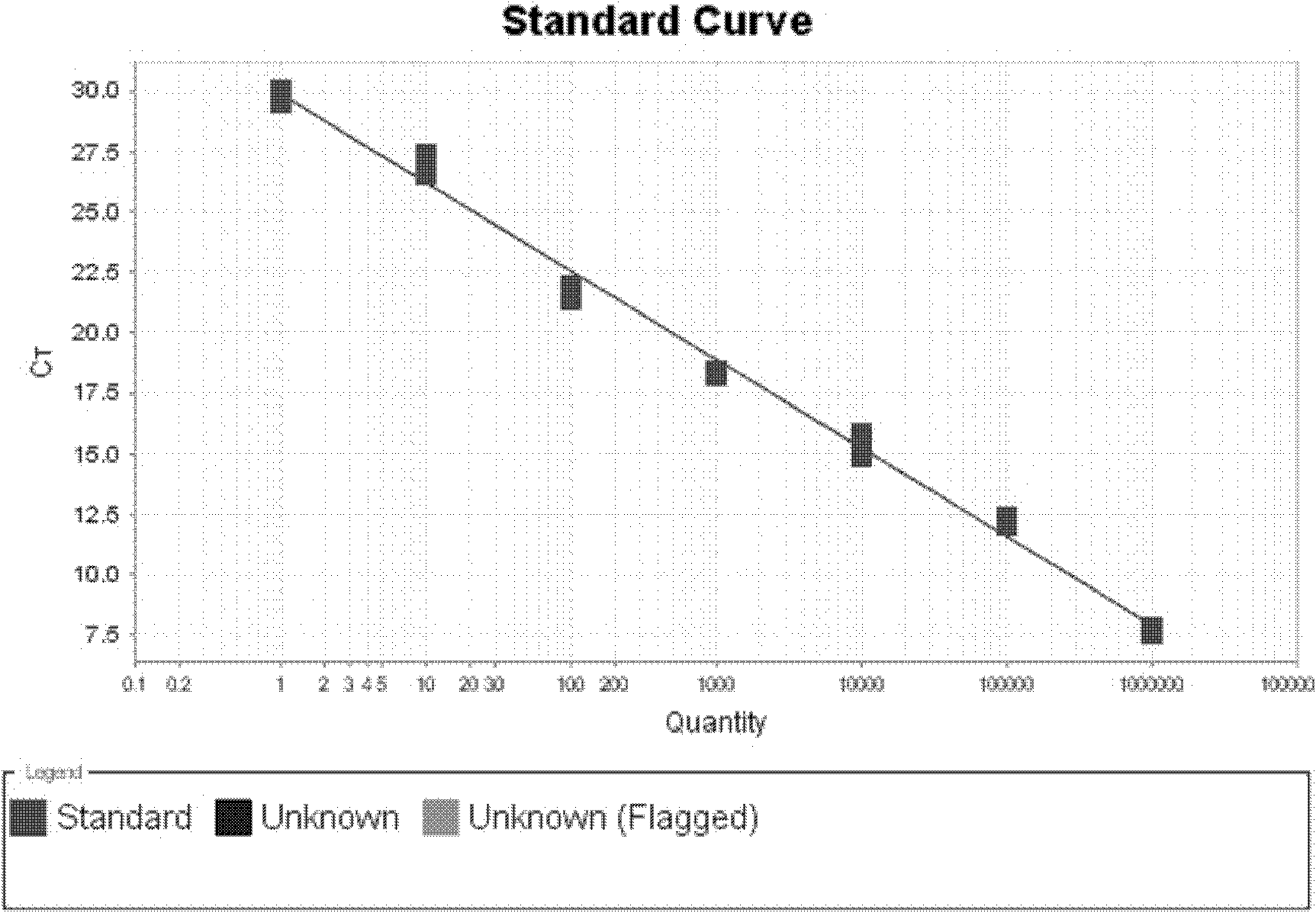
[0089] Example 3: Quantitative determination of Saccharomyces cerevisiae in feed samples
[0090] Quantitative determination of Saccharomyces cerevisiae was carried out on the different samples mentioned in Example 1. Its determination steps are as follows:
[0091] A. Preparation of sample total RNA
[0092] Use enzymatic method to break the yeast cell wall and extract total RNA with high-purity total RNA rapid extraction kit (spin column type): add 0.5-1.0 g of sample to a 1.5 mL centrifuge tube, add 600 μL of sorbitol buffer, add 50 U of Lyticase, and mix well Treat at 30°C for 30 minutes, centrifuge at 1500×g for 10 minutes, discard the supernatant, collect the precipitate, extract the sample RNA according to the instructions of the high-purity total RNA rapid extraction kit, and then treat it twice with DNaseI to ensure that the DNA contamination in the RNA is completely eliminated. For the purified RNA sample, use a UV spectrophotometer to measure the concentration, us...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com