Eukaryotic cell nucleic acid extraction method
A cell nucleic acid and extraction method technology, applied in the field of eukaryotic cell nucleic acid extraction, can solve the problems of few types of samples in the extraction method, poor applicability of large-scale extraction, cumbersome extraction steps, etc., and achieve rapid nucleic acid extraction and wide application , the effect of reducing difficulty and complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment 1, establishment of nucleic acid extraction method
[0030] 1. Extraction solution configuration
[0031] The extraction solution is DEB (DNA extraction buffer) solution;
[0032] DEB solution is an alkaline aqueous solution, the solvent is double distilled water, the solute is an alkaline chemical reagent (such as sodium hydroxide or potassium hydroxide), the configuration method is: take a certain amount of double distilled water, add sodium hydroxide or potassium hydroxide powder , stirring continuously until dissolved, and measuring the pH value, the pH value is 8-13, preferably 10.
[0033] 2. Extraction
[0034] Taking the batch extraction of 96-well plate as an example, the specific operation steps are as follows:
[0035] 1) Add the samples to be extracted into 96-well plates;
[0036] 2) Add a certain amount (such as 120 μL) of DEB (DNA extraction buffer) solution (aqueous solution with a pH between 8-13) to 1), and seal the 96-well plate with 8 ...
Embodiment 2
[0039] Nucleic acid extraction in embodiment 2, whole blood
[0040] One, method of the present invention and Chelex-100 extraction method
[0041] Experimental group: extract according to the extract and method in Example 1:
[0042] 1) Add 2 μL of well-mixed isolated human venous whole blood to a 96-well plate;
[0043] 2) Add 120 μL of DEB solution (sodium hydroxide aqueous solution with a concentration of 0.004 g / L; pH value 10) to 1), the volume ratio of whole blood and DEB is 1 / 60, and seal the 96-well plate with a membrane;
[0044] 3) 100°C boiling water bath for 10 minutes; after cooling, get DEB to extract nucleic acid.
[0045] Control group: Chelex-100 extraction method
[0046] 1) Take 10μL of whole blood and add it to a 1.5mL centrifuge tube;
[0047] 2) Add 500 μL of pure water, shake vigorously, and place at room temperature for 15 minutes;
[0048] 3) Centrifuge at 13,000rpm for 3min, remove the supernatant;
[0049] 4) Repeat the above two steps once to...
Embodiment 3
[0073] Example 3, Nucleic Acid Extraction in Dried Blood Sheets
[0074] One, method of the present invention and Chelex-100 extraction method
[0075] test group:
[0076] 1) Add newborn heel blood dried blood slices (obtained by punching) with a diameter of 3mm into a 96-well plate;
[0077] 2) same as 2) of the experimental group of embodiment 2, (concentration is the sodium hydroxide aqueous solution of 0.004g / L; pH value 10,);
[0078] 3) The same as 3) of the experimental group of Example 2, to obtain alkaline DEB to extract nucleic acid.
[0079] Control group:
[0080] 1) Take a dried blood piece with a diameter of 3mm and add it to a 1.5mL centrifuge tube;
[0081] 2)-8) are the same as 2)-8) of the control group in Example 2 to obtain Chelex-100 extracted nucleic acid.
[0082] 2. Comparison of two methods for extracting nucleic acid
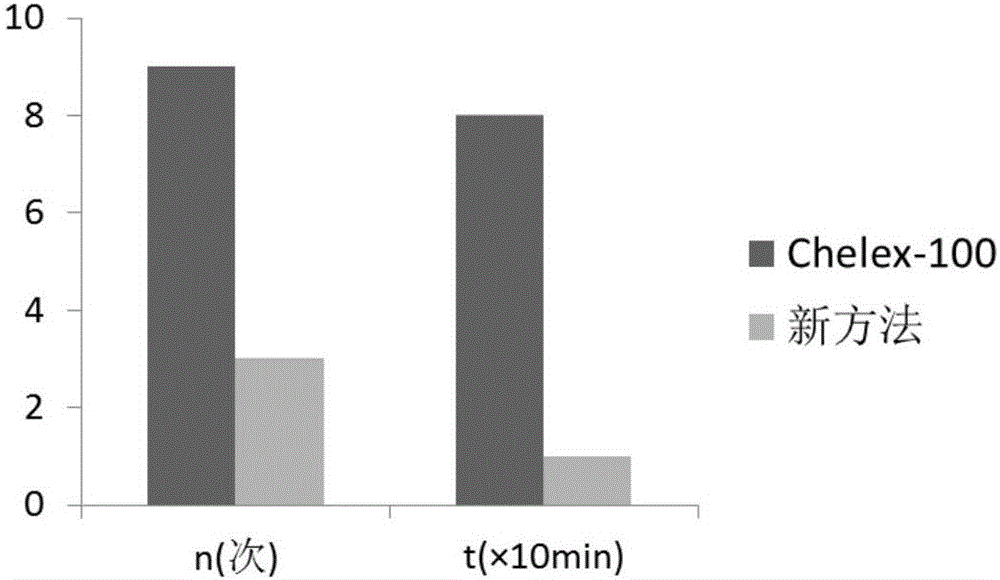
[0083] 1. Number of steps and time required for extraction
[0084] Same as embodiment 2;
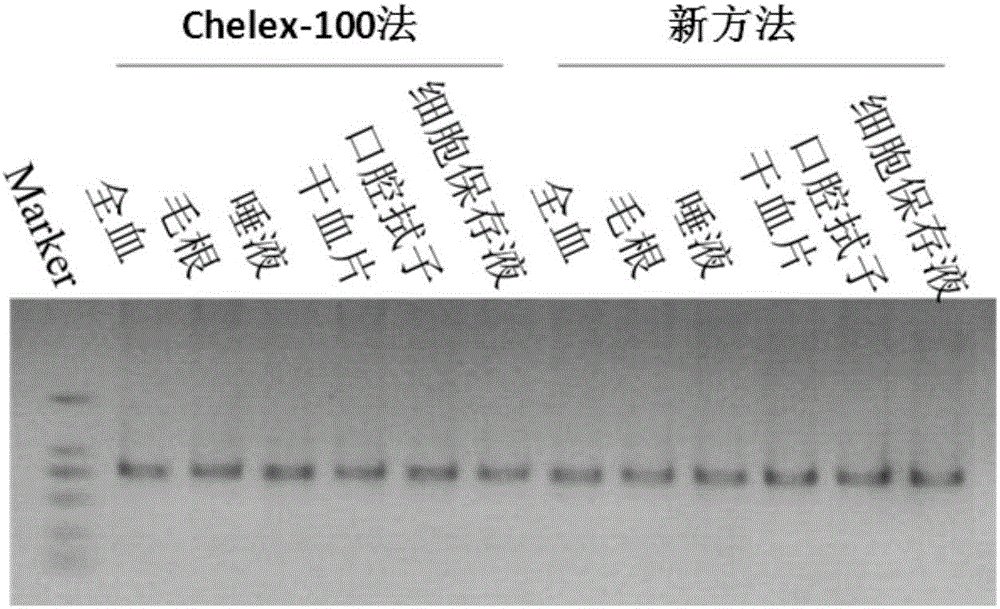
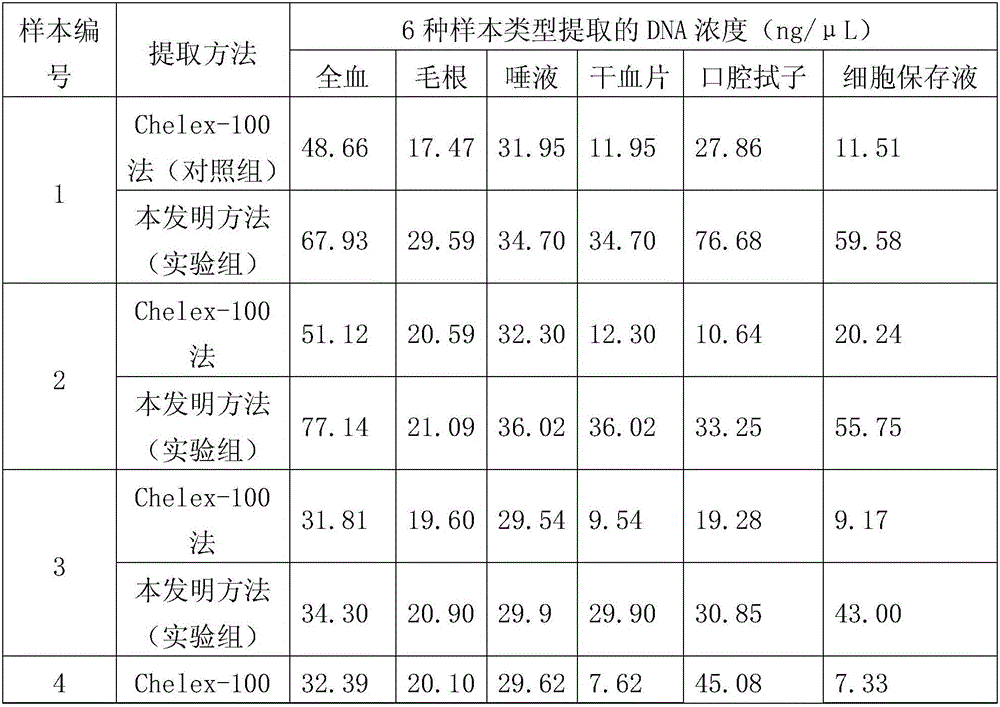
[0085] 2. Nucleic acid extraction...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com