Nested PCR (polymerase chain reaction) detection primer and detection method for Uromyces vignae
A technology for detecting primers and single cells, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: Design of specific primers for PCR detection of P. cowpea and verification of specificity of primers
[0031]1. Extraction of Genomic DNA from Monocystis cowpea and other strains
[0032] The CTAB method was used to extract the genomic DNA of Monocystis cowpea and other bacterial strains. The specific method was as follows: the sample to be tested was ground into powder in liquid nitrogen, and 100 μg of the powdered sample to be tested was put into a 2 mL centrifuge tube, and 900 µL 2% CTAB (cetyltrimethylammonium bromide) extract (2% CTAB; 100 mmol / L Tris-HCl, pH=8.0; 20 mmol / L EDTA, pH=8.0; 1.4 mol / L NaCl) and 20-30 μL β-mercaptoethanol [Note: CTAB and β-mercaptoethanol need to be mixed at 65 °C], use a shaker to shake and mix, 65 °C water bath for 30 min (shake the centrifuge tube 5-6 times in the middle, (to fully release the DNA into the buffer), take out the tube, cool to room temperature, add 300 μL phenol to mix evenly, let it stand at room temperatu...
Embodiment 2
[0041] Embodiment 2: Sensitivity determination of cowpea monocell rust nested PCR detection
[0042] 1. Routine PCR detection
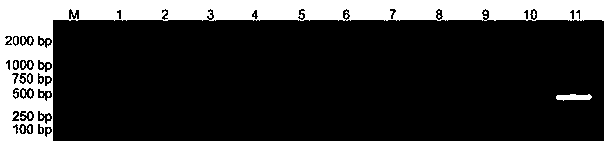
[0043] Dilute the genomic DNA of Monocystis vignaceae with sterile ultrapure water, and prepare a series of concentrations of 10 times the order of magnitude for future use. Use UV-ITSF / UV-ITSR to perform PCR amplification on genomic DNA with different concentrations, and evaluate the sensitivity of the primers to the detection of the genomic DNA of Monocys vignaceae. The amplification reaction system and reaction procedure are as follows: PCR reaction system 10 µL , including 5 µL of 2×Taq PCR Master Mix (Beijing Tiangen Biochemical Technology Co., Ltd.), 0.5 µL of 10 nmol / L UV-ITSF / UV-ITSR primers, 1.0 µL of DNA template, and make up to 10 µL. The amplification reaction program was: pre-denaturation at 94 °C for 4 min; 30 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 7...
Embodiment 3
[0049] Example 3: Nested PCR detection of Adzuki bean leaf rust at different times after inoculation
[0050] Obtaining of diseased tissues by artificial inoculation: select Baoqinghong seeds with full grains and uniform size, accelerate germination in a constant temperature incubator at 25 °C, and sow them in flowerpots with a diameter of 15 cm after the seeds germinate, sow 9 seeds in each pot, and sow Afterwards, they were cultured in a greenhouse under the conditions of 16 h light (25 °C) / 8 h dark (20 °C). When the true leaf expansion was 70-80%, the true leaves of adzuki bean were inoculated by referring to the reported method, and Samples were taken at 0 h, 12 h, 24 h, 48 h, 120 h, and 192 h after inoculation. In order to remove the uredia spores remaining on the surface of the leaves during inoculation, the leaves of each sample were washed with tap water on both sides. Rinse again with sterile water for 2-3 times. After rinsing, blot the surface moisture with sterilize...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com