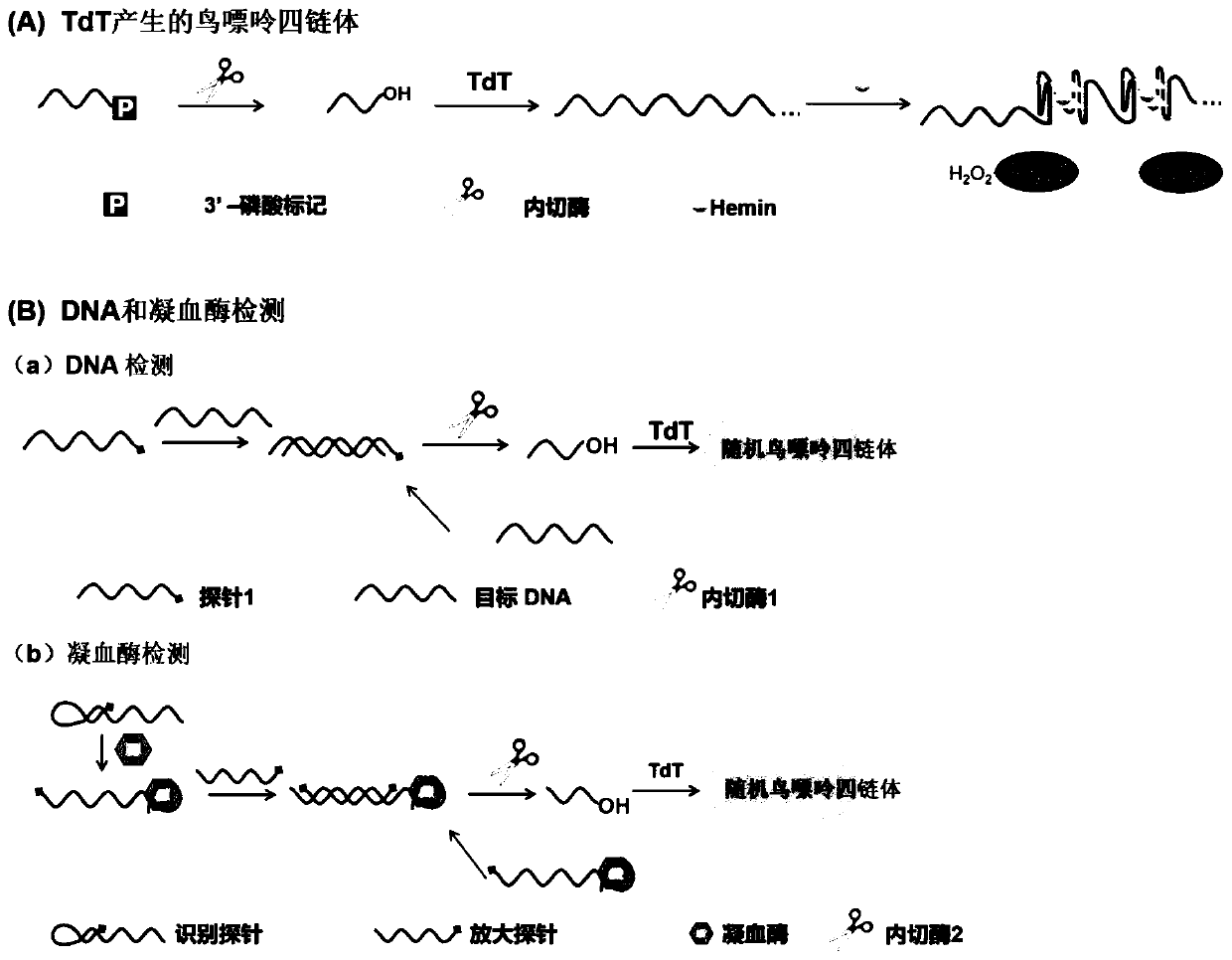
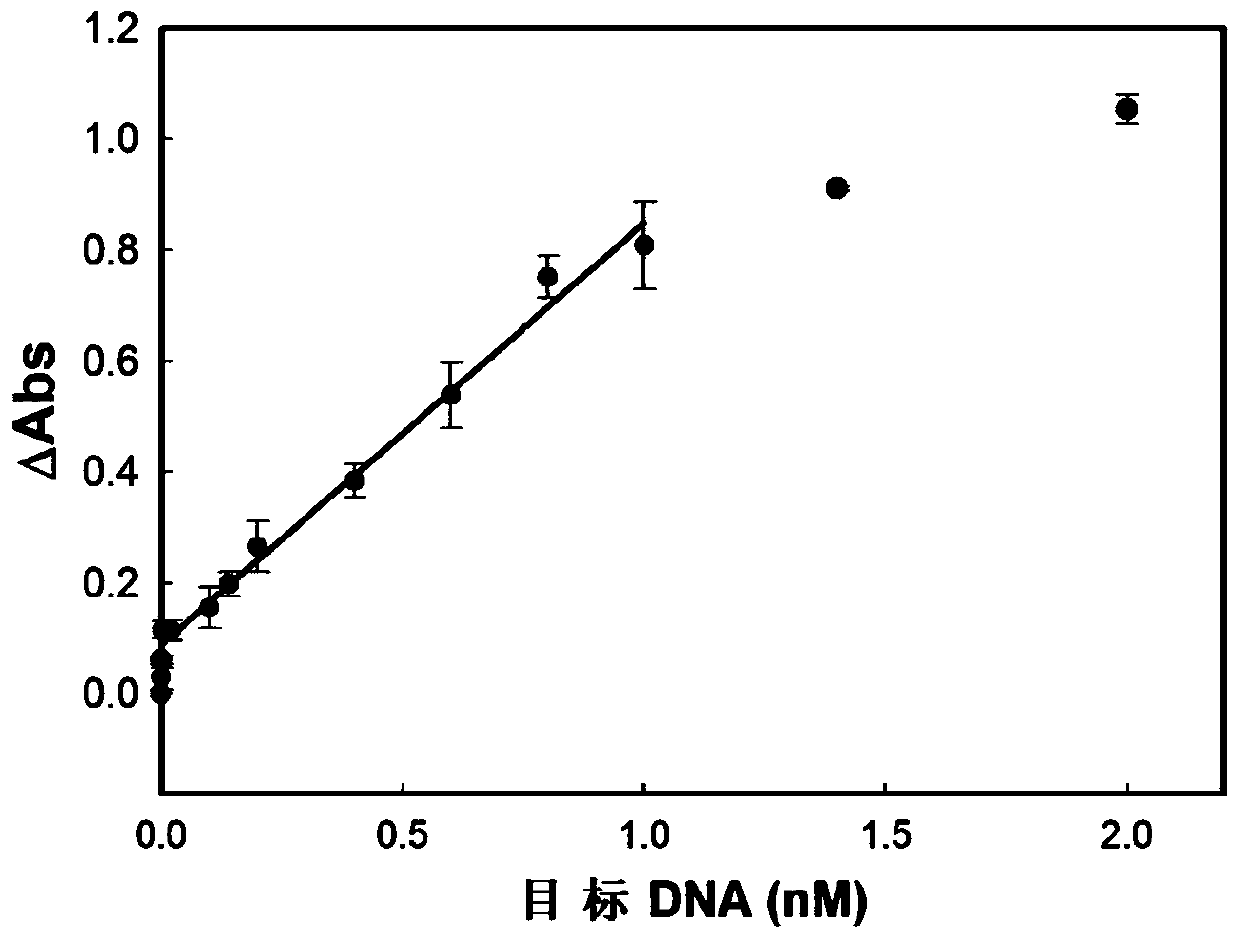
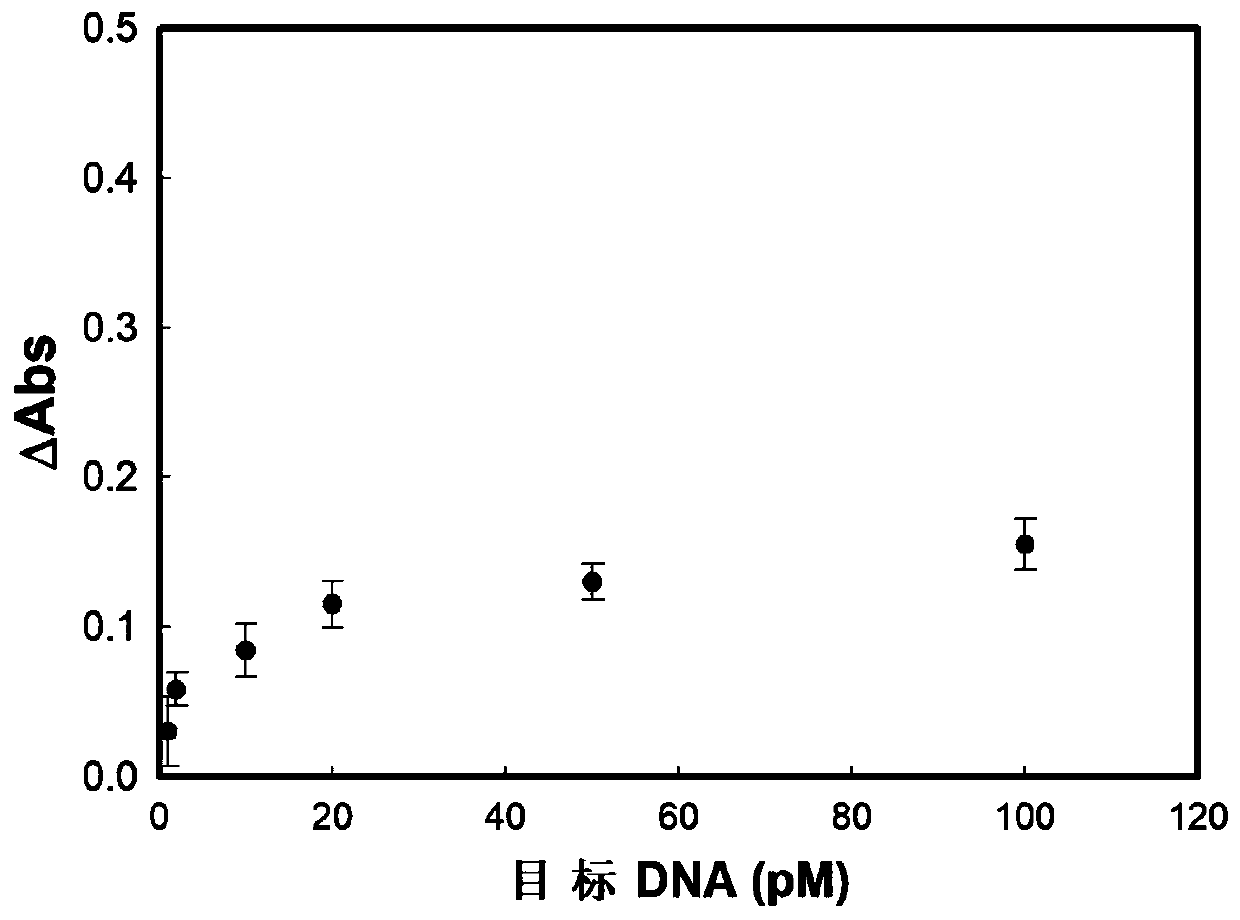
Method for detecting DNA or protein based on double-amplification method
A protein and dual technology, applied in biochemical equipment and methods, measurement devices, and microbial assay/inspection, etc., can solve the problems of incomplete guanine quadruplex sequence sealing, increased design difficulty, high background signal, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Embodiment 1 detects target DNA
[0059] 1. Target identification
[0060] Recognition probe 5'-AAA GGA TCG AAT AAG AGG TCC TTC-PO 4 3- 3' (SEQ ID NO. 1);
[0061] Target nucleic acid 5'-GAA GGA CCT CTT ATT CGATCC TTT-3' (SEQ ID NO.2);
[0062] In a 600 μL centrifuge tube, add 1 μL of 15 μM recognition probe, 1 μL of 100 nM target nucleic acid, 2 μL of 5×NBE buffer 4 (100 mM Tris-Ac, 50 mM Mg(Ac) 2 , 250mM KAc, pH 7.9) buffer solution, and 6 μL ultrapure water, mixed with a 10 μL pipette gun, and incubated in a 58°C water bath for 5 minutes.
[0063] 2. Endonuclease amplification detection
[0064] After 5 minutes, add 8 units of Nt.AlwI to the centrifuge tube and incubate at this temperature for 2 hours. Add the endonuclease and incubate at 37°C for 2 hours; during incubation, the endonuclease will cut the probe and release the DNA fragment containing 3'OH 5'-AAA GGA TCG AAT-OH-3'(SEQ ID NO.3). After 2 hours, the nicking reaction was terminated by incubating th...
Embodiment 2
[0069] Embodiment 2 detects thrombin
[0070] 1. Target identification
[0071] The identification probes are:
[0072] 5'-AGTCCGTGGTAGGGCAGGTTGGGGTGACTGCTGATTTTTGAGCCTCAGCAGTCACCC-PO 4 3- 3' (SEQ ID NO.4);
[0073] The amplification probe is: 5'-TTTTTAGACTGCTGAGGC-PO 4 3 -3' (SEQ ID NO.5);
[0074] The specific sequences of the recognition probe and amplification probe are designed by the applicant according to the characteristics of thrombin itself. The recognition probe includes three parts, AGTCCGTGGTAGGGCAGGTTGGGGTGACT (5'-3') is a thrombin nucleic acid aptamer sequence, which can recognize and bind thrombin. The second part is the restriction endonuclease nicking site of CCTCAGC (5'-3'), which is used as the signal initiation part. The third part is the hairpin structure formed after the recognition probe is annealed, which contains a 13-base complementary stem, which acts as a closed hairpin. The sequence length of the amplified probe is not more than 20bp, and t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com