Identifying and breeding method of SNP marker for increasing cotton fiber yield and high-yield cotton
A technology of marking and cotton, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, recombinant DNA technology, etc., can solve the problems of being easily affected by the environment, time-consuming and labor-consuming, and low selection efficiency, so as to improve selection efficiency and Accuracy, the effect of shortening the cultivation period
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
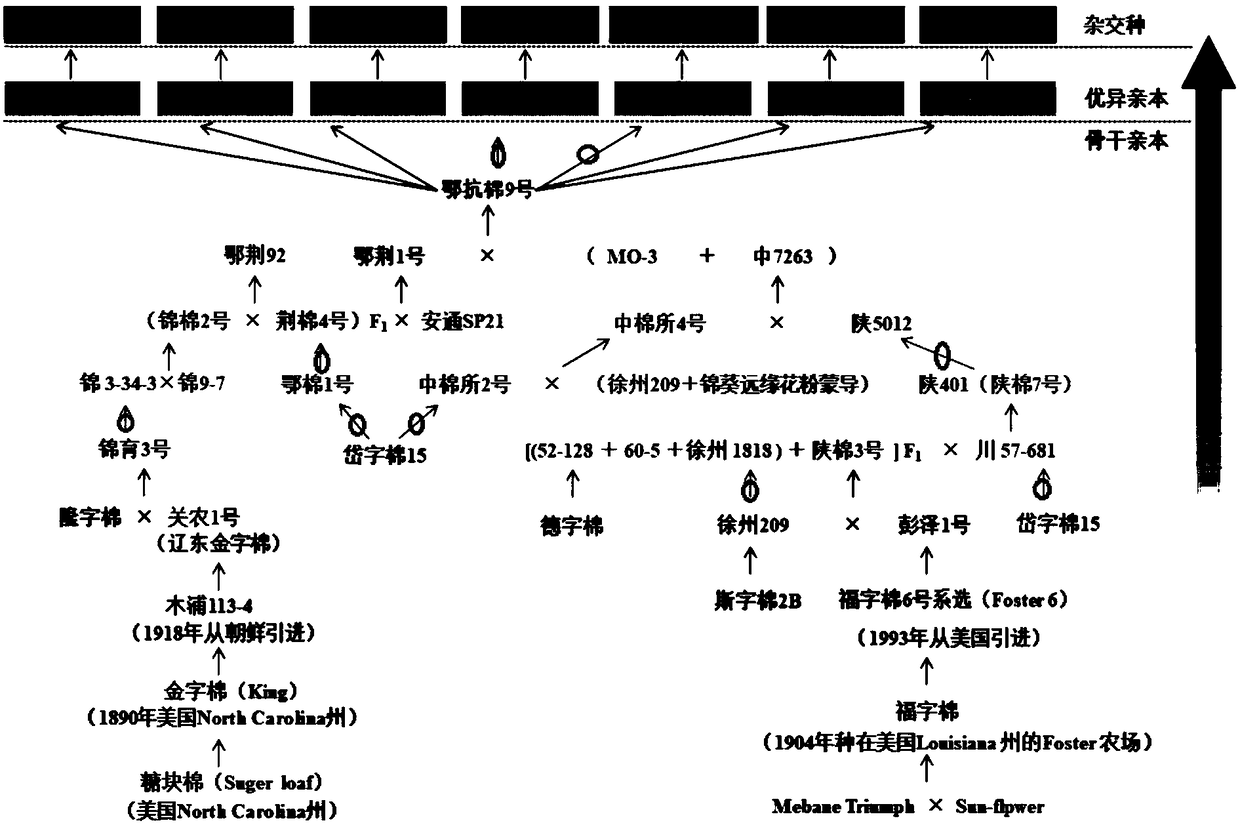
[0047] 1. Tracking of the pedigree of the female parent of strong dominant hybrid cotton
[0048] The breeding and improvement process of Ekangmian 9 was sorted out, and the complete pedigree improvement information from the introduction of foreign species was obtained ( figure 1 ). During the improvement of the pedigree, the genetic components of the upland cotton lines Jinzi cotton, Daizi cotton 15, Longzi cotton, Dezi cotton, Sizi cotton 2B and Fuzi cotton were collected, and the yield, quality, resistance and environmental adaptation were realized. Synchronized improvement of sex.
[0049] 2. The ecological adaptation improvement process of the Yangtze River Basin
[0050]According to our tracking of the parent breeding process of excellent hybrids, we found that the adaptability of these excellent hybrids to the Yangtze River Basin was mainly inherited from Ejing 1. The earliest origin of Ejing 1 can be traced to sugar loaf in North Carolina (NC), USA. In 1890, King T...
Embodiment 2
[0062] Genealogical Core Material Whole Genome Resequencing
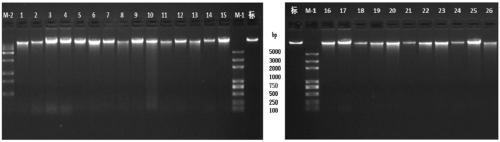
[0063] 1. Genomic DNA extraction and quality testing
[0064] The whole genome DNA (Paterson et al., 1993) of 26 test materials (see Table 1 in Example 1) was extracted by the improved CTAB method, and the samples were detected by agarose electrophoresis, Nanodrop microspectrophotometer and Qubit Fluorometer fluorescence quantitative instrument Purity, integrity, concentration, and DNA quality test results showed that all sample DNA met the requirements for library construction and sequencing. The test results are as follows:
[0065] 1.1 DNA purity and integrity test results
[0066] Agarose electrophoresis: agarose concentration: 1%; voltage: 100v; electrophoresis time: 40min. The result is as image 3 shown.
[0067] 1.2 Purity and concentration test results
[0068] For the samples with qualified agarose detection results, the purity and concentration of the samples were further detected by Nanodrop micro-...
Embodiment 3
[0079] On the basis of 26 pedigree materials, the deep sequencing data of Sizimian 2B was further collected for analysis (SRR5512449) (Fang L et al., 2017). Using 27 family materials as cultivar groups, according to the SNP detection method, the SNP variation in the 27 materials was detected to obtain the population SNP variation. At the same time, the sequencing data of 31 wild cotton varieties were downloaded from the database (Wang M et al., 2017). The SNP variations detected in the above 27 family varieties and 31 wild cotton were used to construct the population phylogenetic tree and analyze the selection and elimination.
[0080] 1. Experimental method
[0081] 1.1 SNP detection method
[0082] SAMtools was used to detect the SNP of the test materials. It mainly includes the following steps:
[0083](1) Prepare the reference genome and its index file: use the faix command in the SAMtools software to establish the reference genome index;
[0084] (2) SNP detection: u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com