Method utilizing liquid phase capture to carry out gene typing of large yellow croaker genome
A technology of genotyping and large yellow croaker, applied in biochemical equipment and methods, microbiological measurement/inspection, etc., can solve the problems of high cost, difficulty in application, and complicated gene chip design, so as to improve efficiency, specificity, Effects of High Efficiency Genome Typing Technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
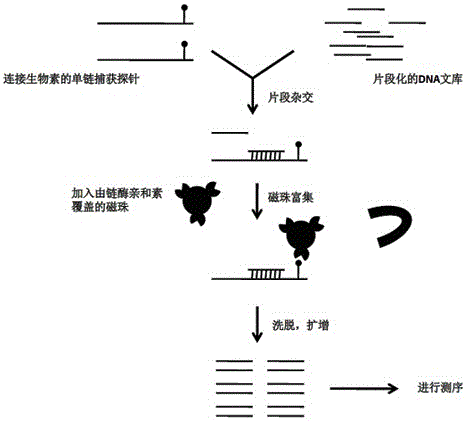
[0023] In the embodiment of the present invention, a liquid-phase capture method for whole-genome genotyping of large yellow croaker:
[0024] 1. Resequencing data using large yellow croaker
[0025] In view of the fact that the above-mentioned individuals used for genome sequencing are highly homozygous individuals with few heterozygous sites / SNP sites, 100 large yellow croakers (50 male and 50 male) were selected from different breeding groups in September 2012 , Genome resequencing was carried out by constructing a 300bp small fragment library. The two groups of resequencing obtained a total of 20Gb of raw data, and the average sequencing depth reached 26.8.
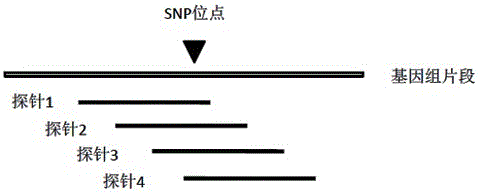
[0026] 2. Determine the capture site, probe design
[0027] The sequencing results were compared to the above draft genome for SNP marker mining. A total of 12,161,573 SNP sites with a sequencing depth greater than 5X, 505,286 on exons, 4,949,058 on introns, and 5,135,488 between genes were obtained. According to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com