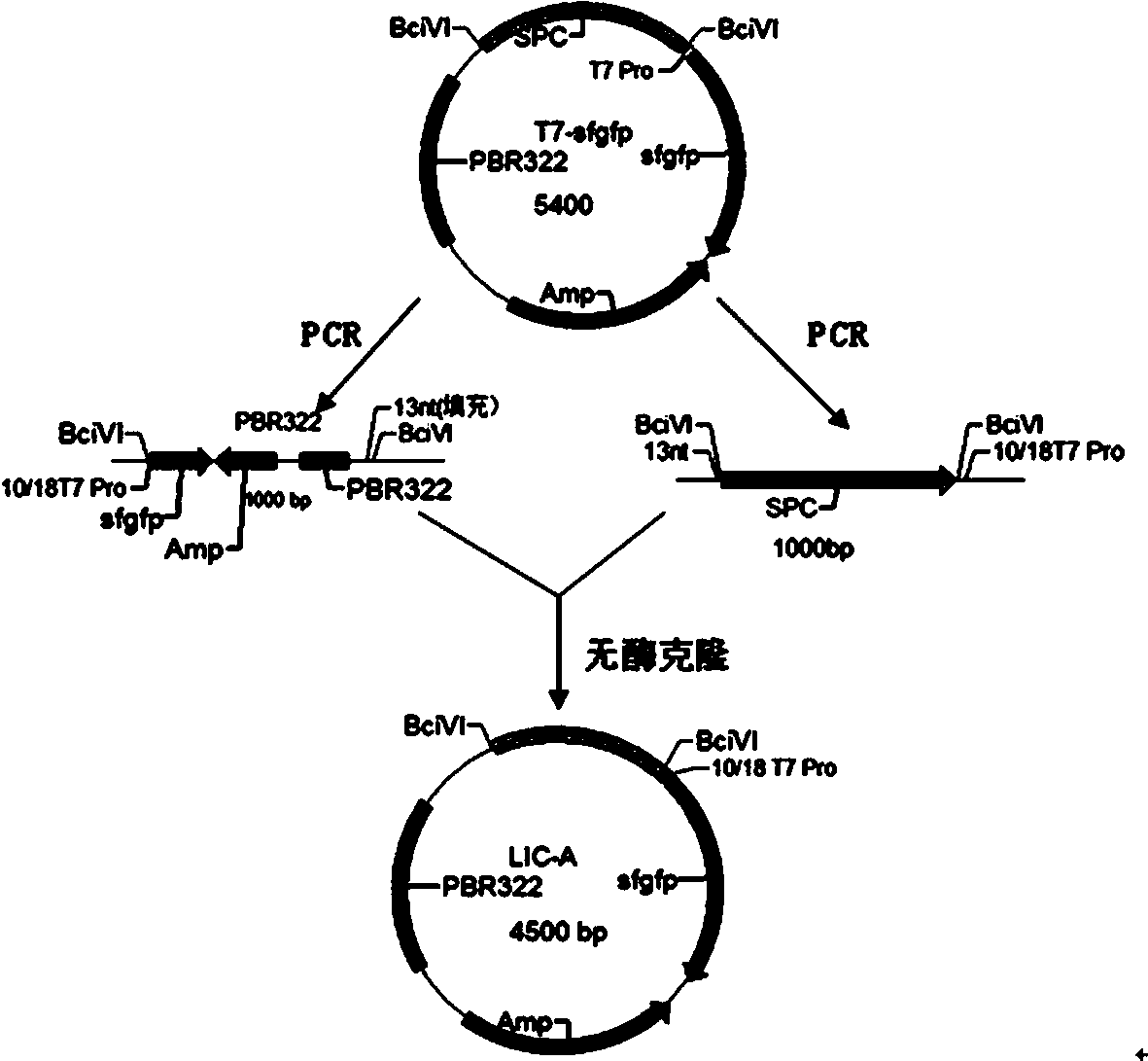
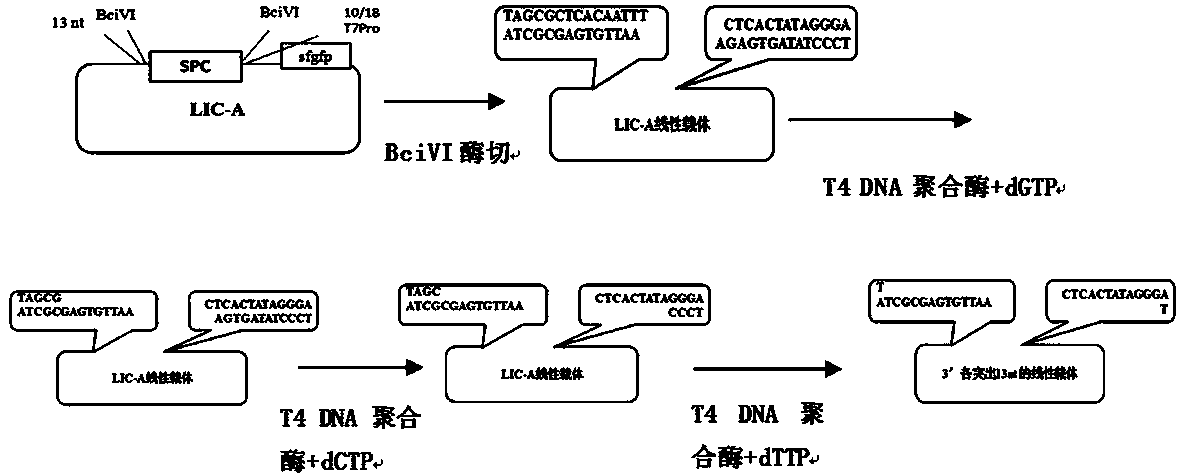
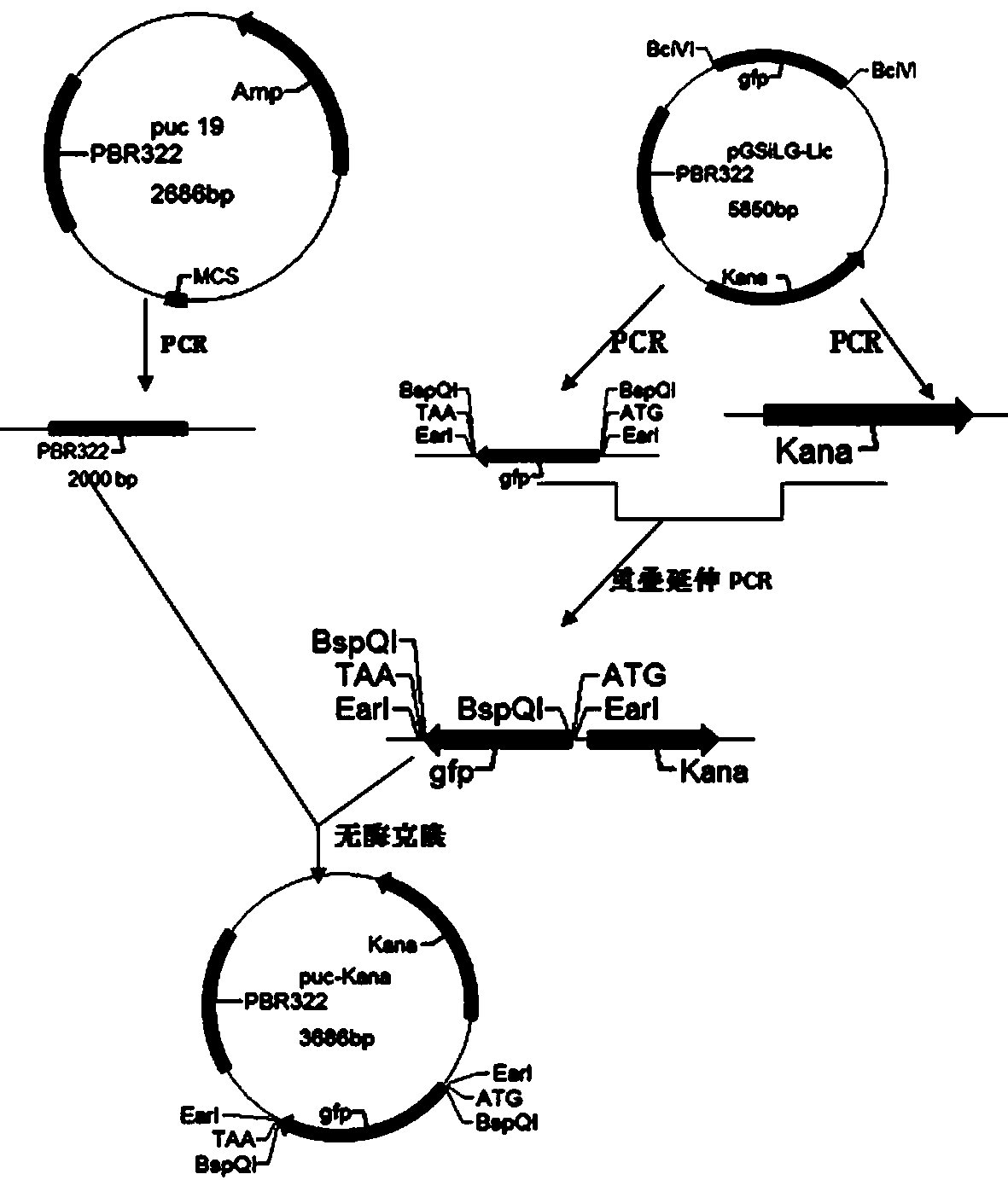
Method for synthesizing DNA fragments and assembling synthetic genes in Escherichia coli through one-step method
A technology of Escherichia coli and gene synthesis, applied in the field of genetic engineering, can solve the problems of high error rate of synthesis method, not very practical research, slow synthesis speed, etc., and achieve the effects of saving experimental reagents, intuitive screening method, and low synthesis cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Using the present invention, a glucose oxidase (AGOX) gene derived from Aspergillus niger was synthesized.
[0034] 1. Use the steps 1), 2) and 3) of the content of the invention to prepare and synthesize LIC-A vector plasmid, LIC-A linear vector, and acceptor vector puc-Kana, respectively, for use.
[0035] 2. First divide the amino acid sequence (589 aa) of the target gene AGOX into 10 small segments, named AG1, AG2,...AG10, each segment is about 55-60 aa, and then design seamlessly in the DNAworks online software Primer sequence, synthesis of 10 sets of primers (each set of 10-12).
[0036] 3. Dilute the synthesized 12 oligonucleotide sequences to a final concentration of 10μM respectively, then take 0.5μL into a PCR tube, add 2ul LA Taq Buffer, and add double distilled water to a final volume of 20μL ( The final concentration is about 0.25μM). During annealing, process the mixture on the PCR machine according to the gradual cooling procedure of 94°C for 5min, 9...
Embodiment 2
[0044] Example 2: Using the present invention, a proline endopeptidase (SCP) gene derived from Penicillium chrysogenum Wisconsin 54-125 was synthesized.
[0045] 1. Use the steps 1), 2) and 3) of the content of the invention to prepare and synthesize LIC-A vector plasmid, LIC-A linear vector, and acceptor vector puc-Kana, respectively, for use.
[0046] 2. First divide the amino acid sequence (543 aa) of the target gene SCP into 9 small segments, named SP1, SP2,...SP9, each segment is about 55-60 aa, and then design seamlessly in the DNAworks online software Primer sequence, synthesis of 9 sets of primers (each set of 10-12).
[0047] 3. Dilute the synthesized 12 oligonucleotide sequences to a final concentration of 10μM respectively, then take 0.5μL into a PCR tube, add 2ul LA Taq Buffer, and add double distilled water to a final volume of 20μL ( The final concentration is about 0.25μM). During annealing, process the mixture on the PCR machine according to the gradual cooling proce...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com