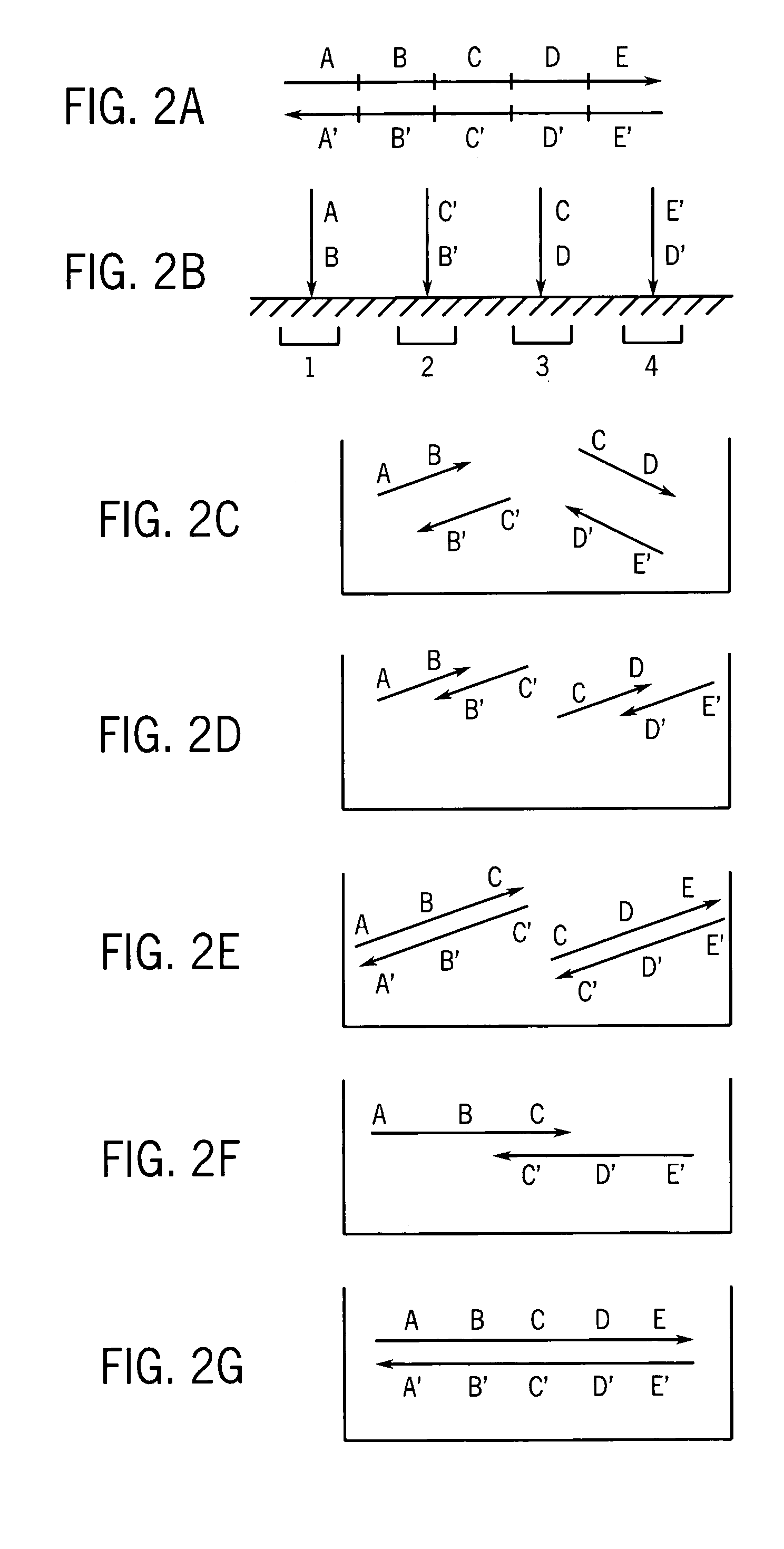
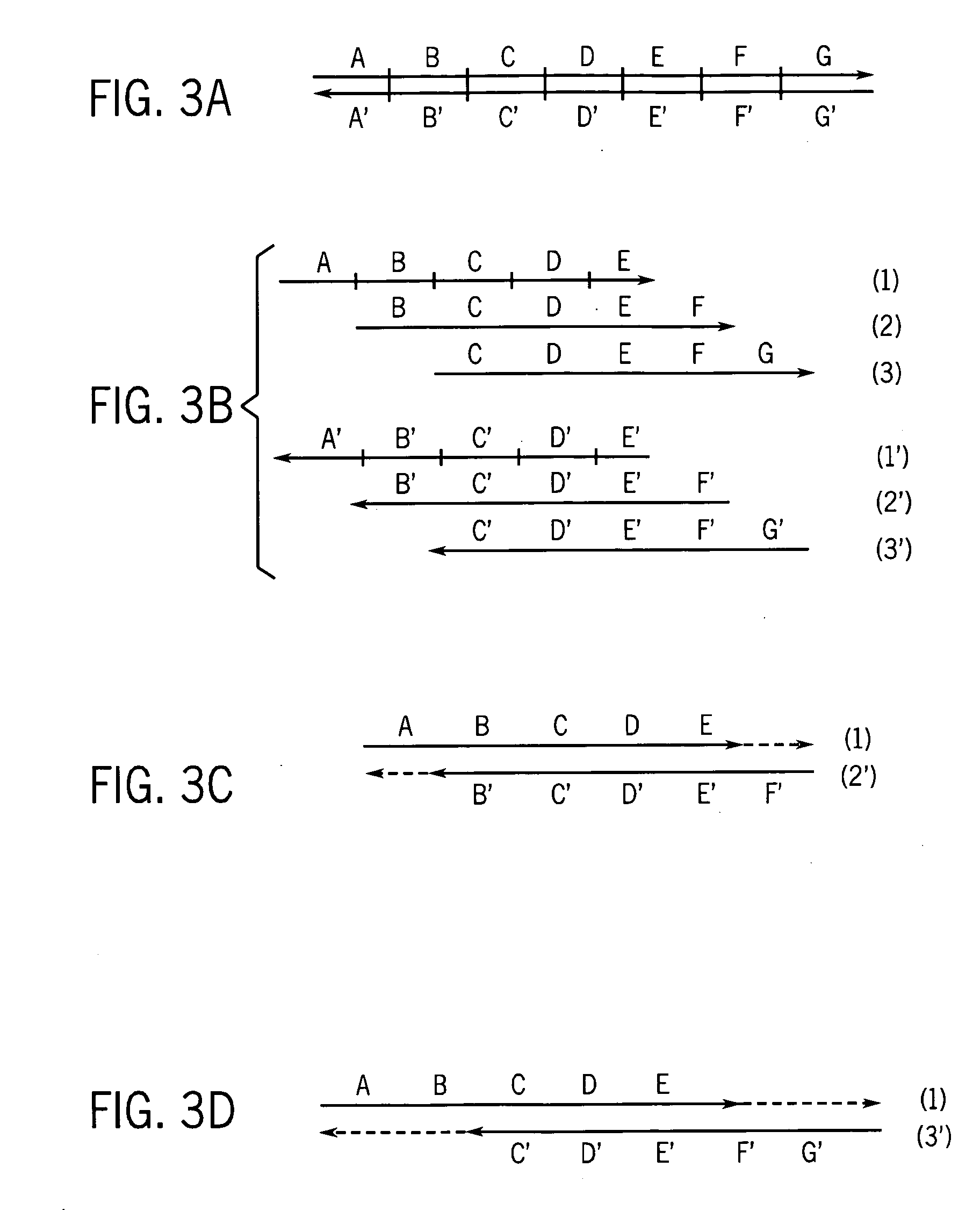
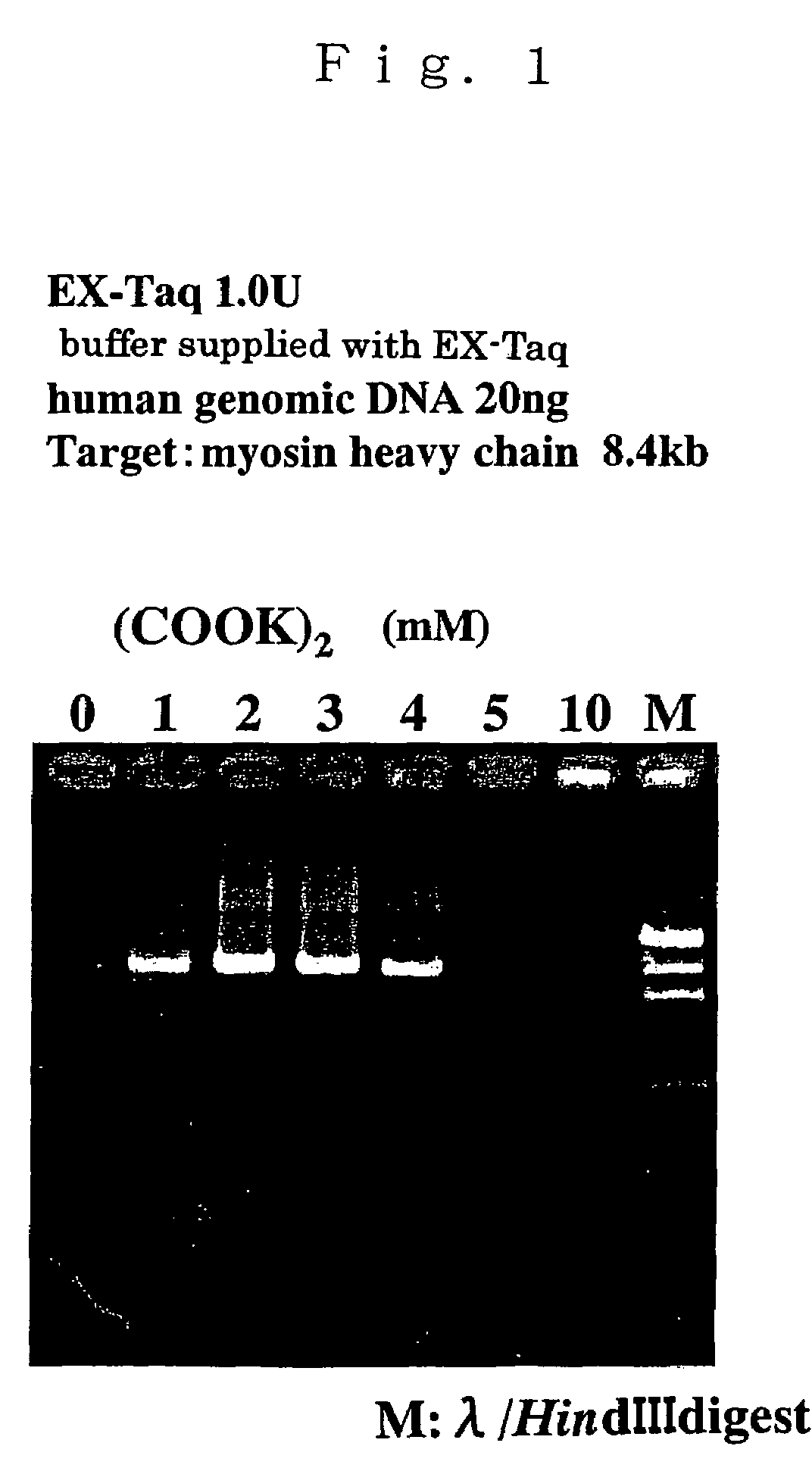
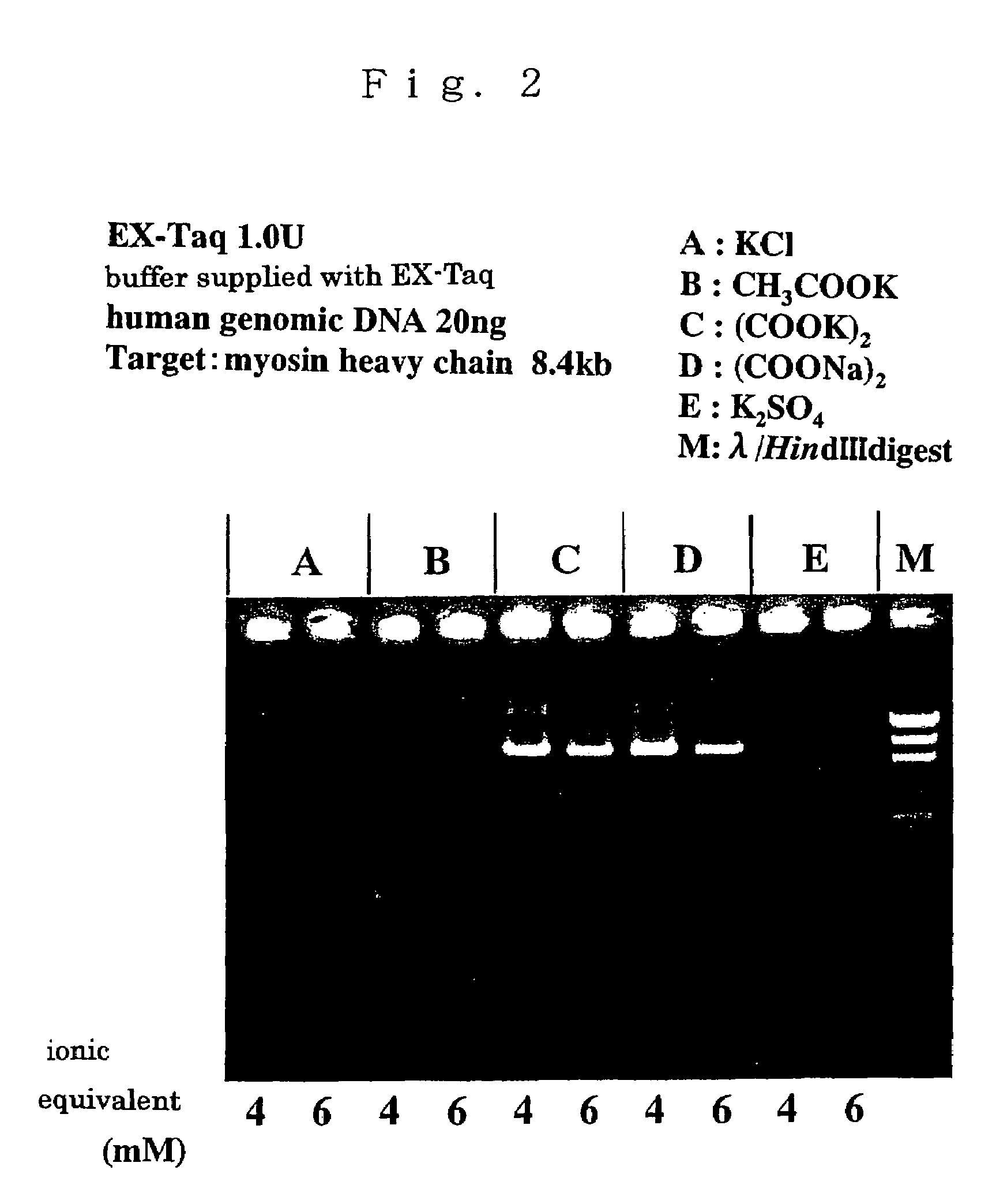
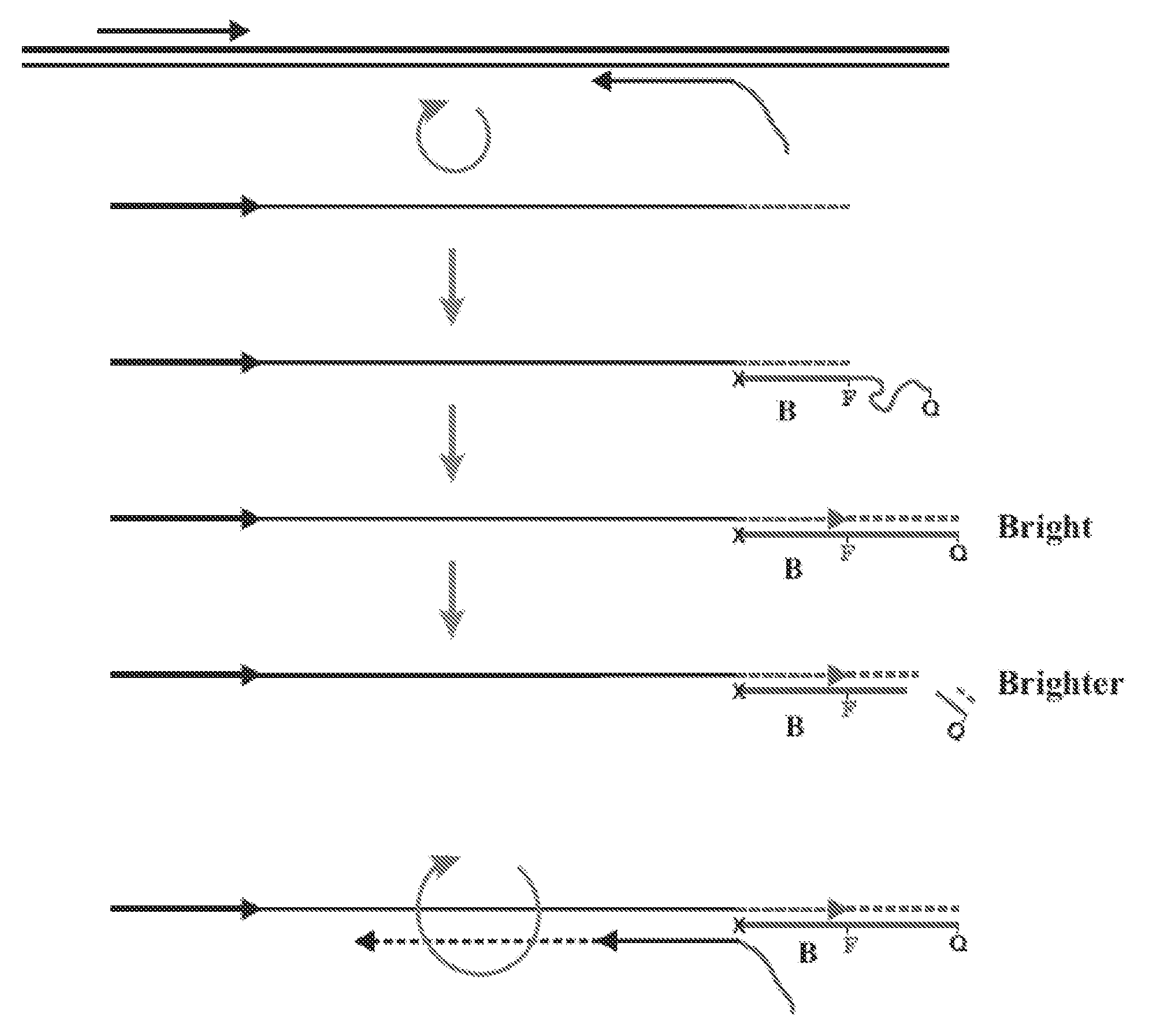
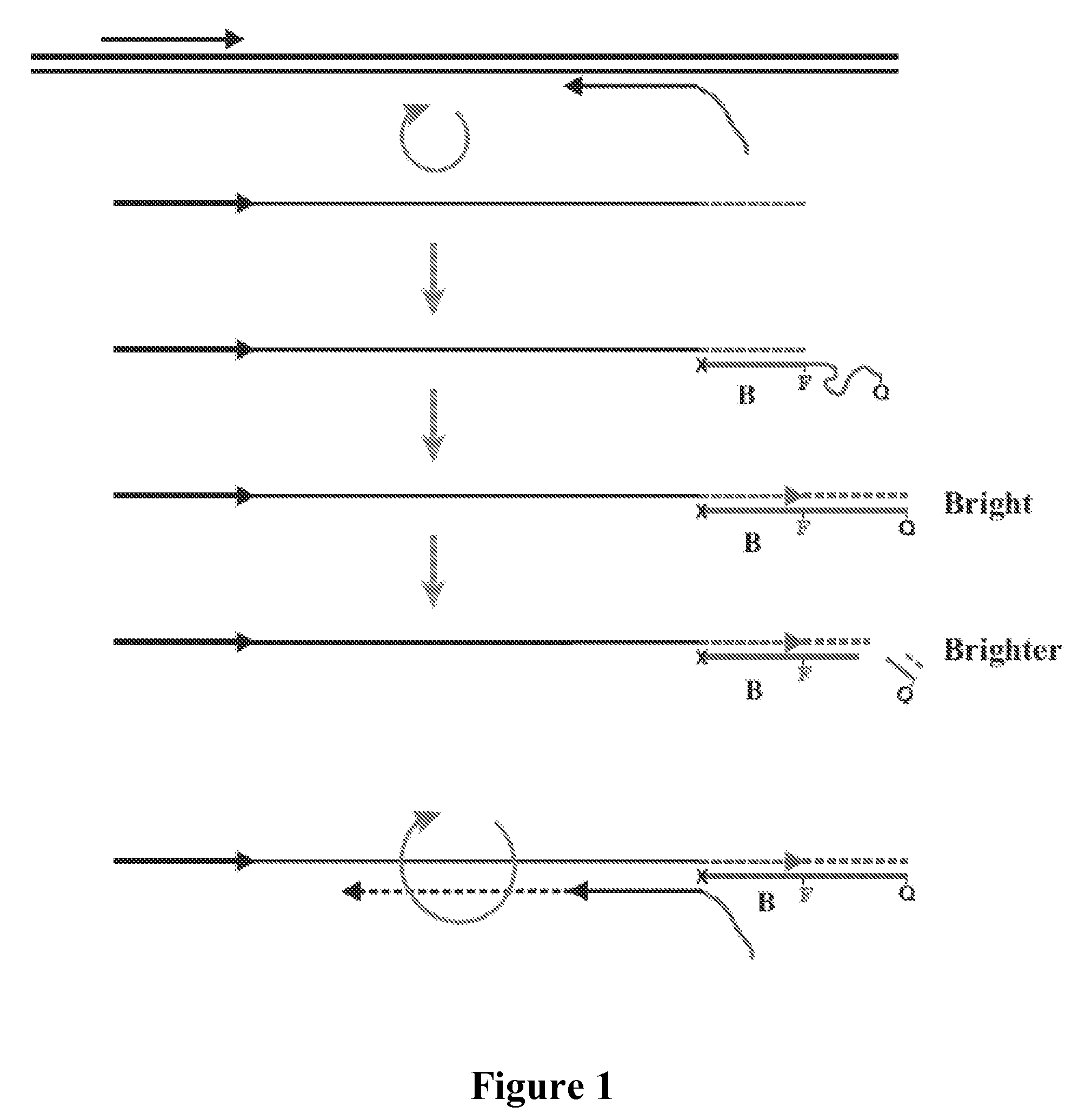
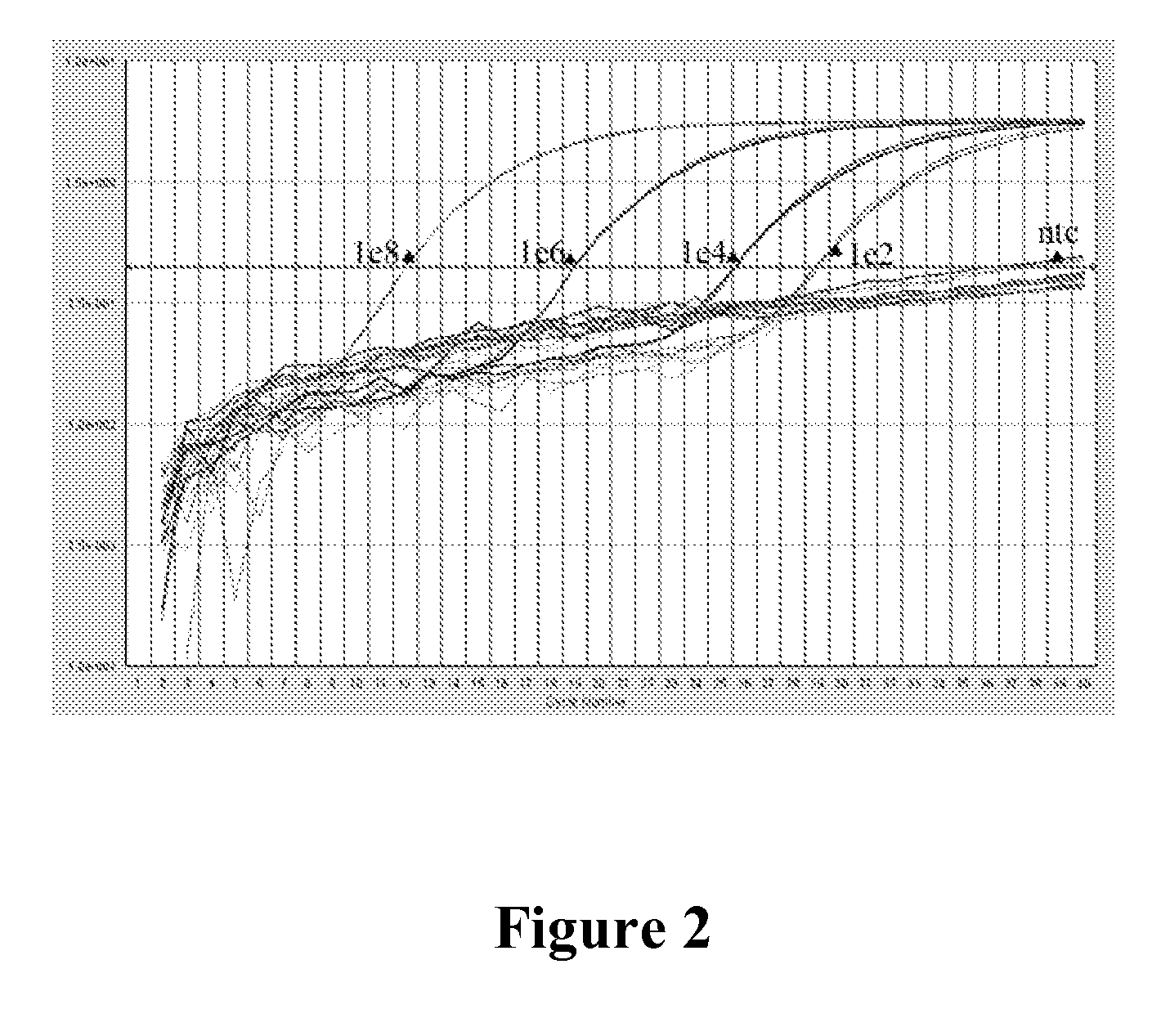
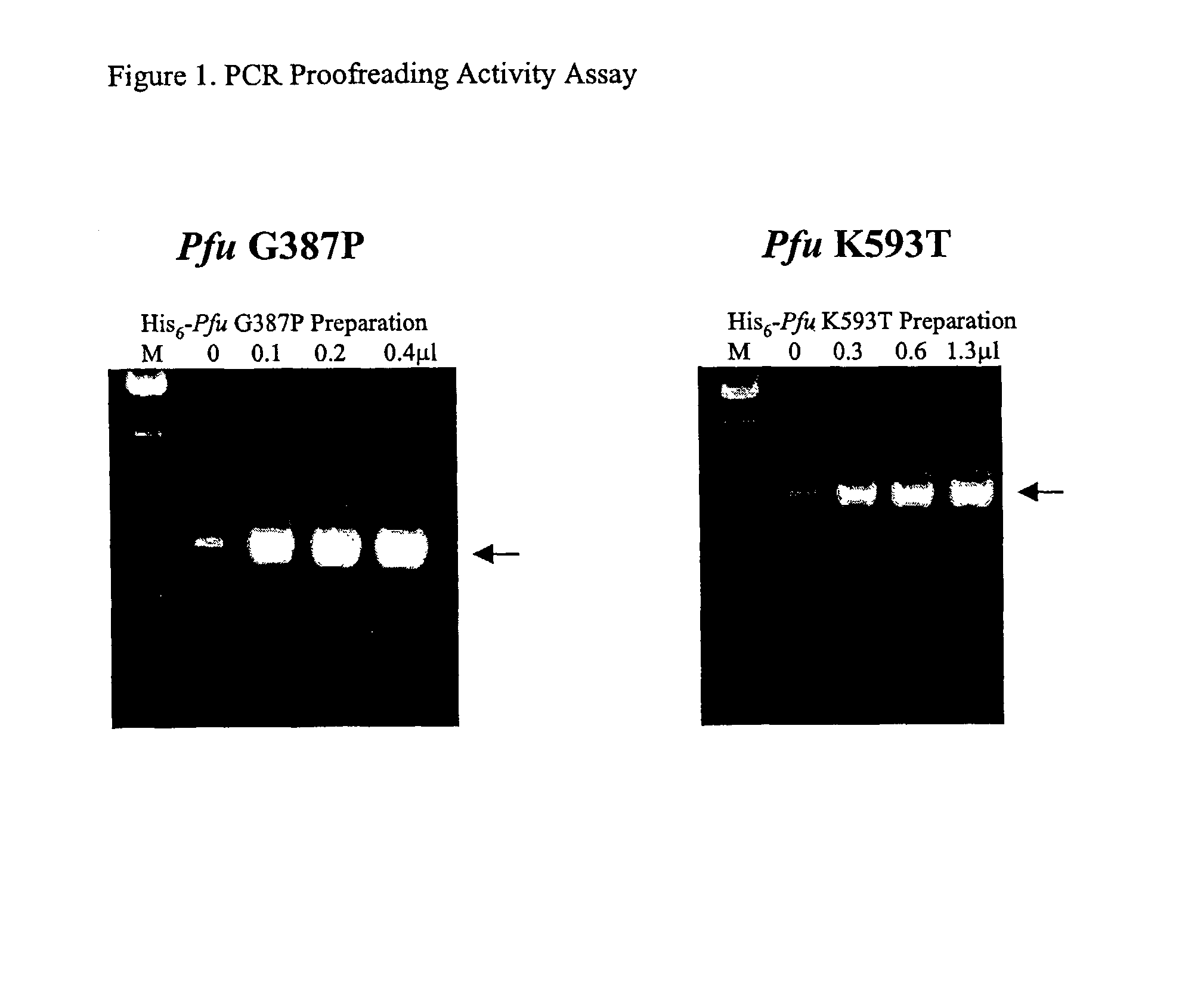
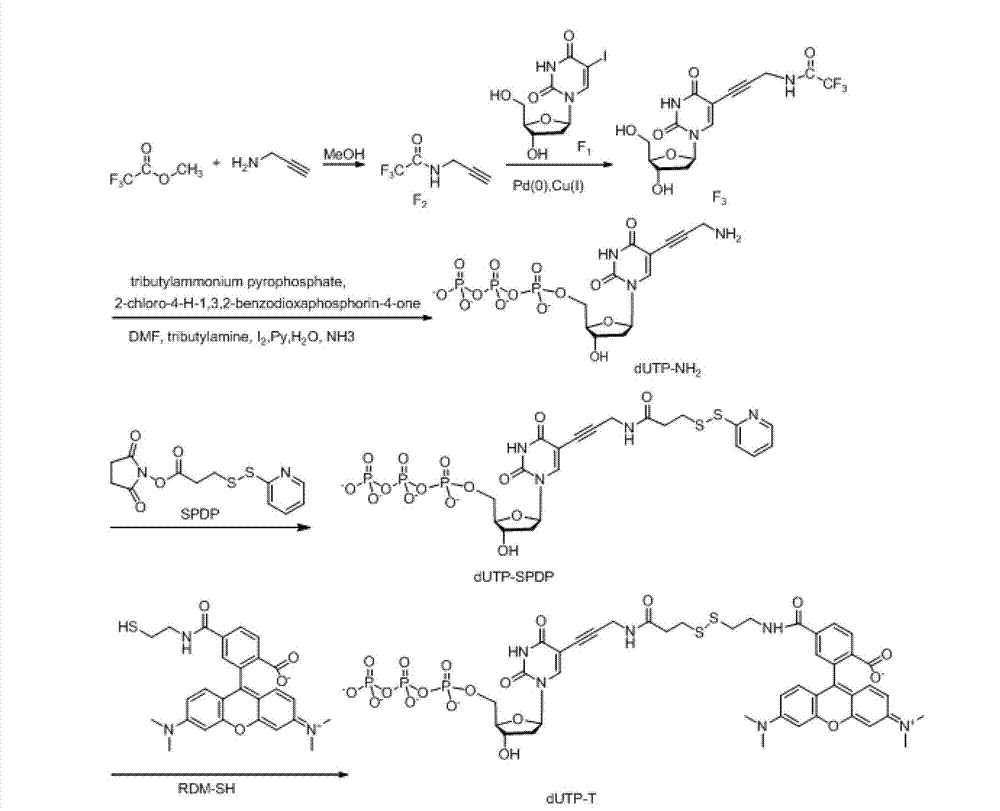
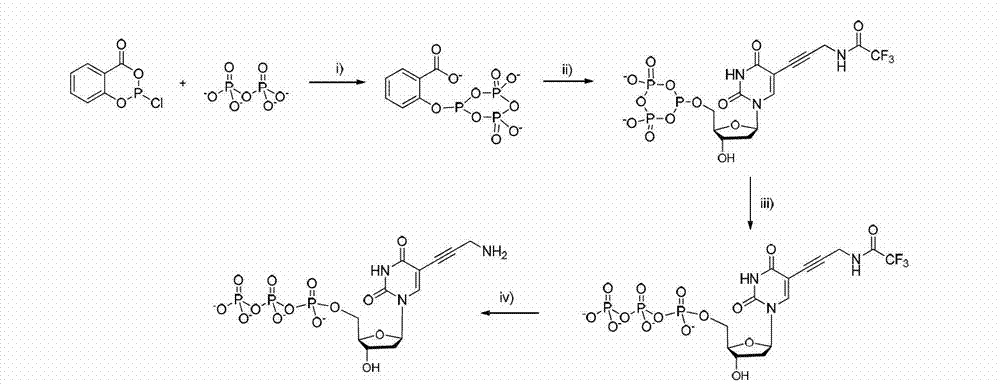
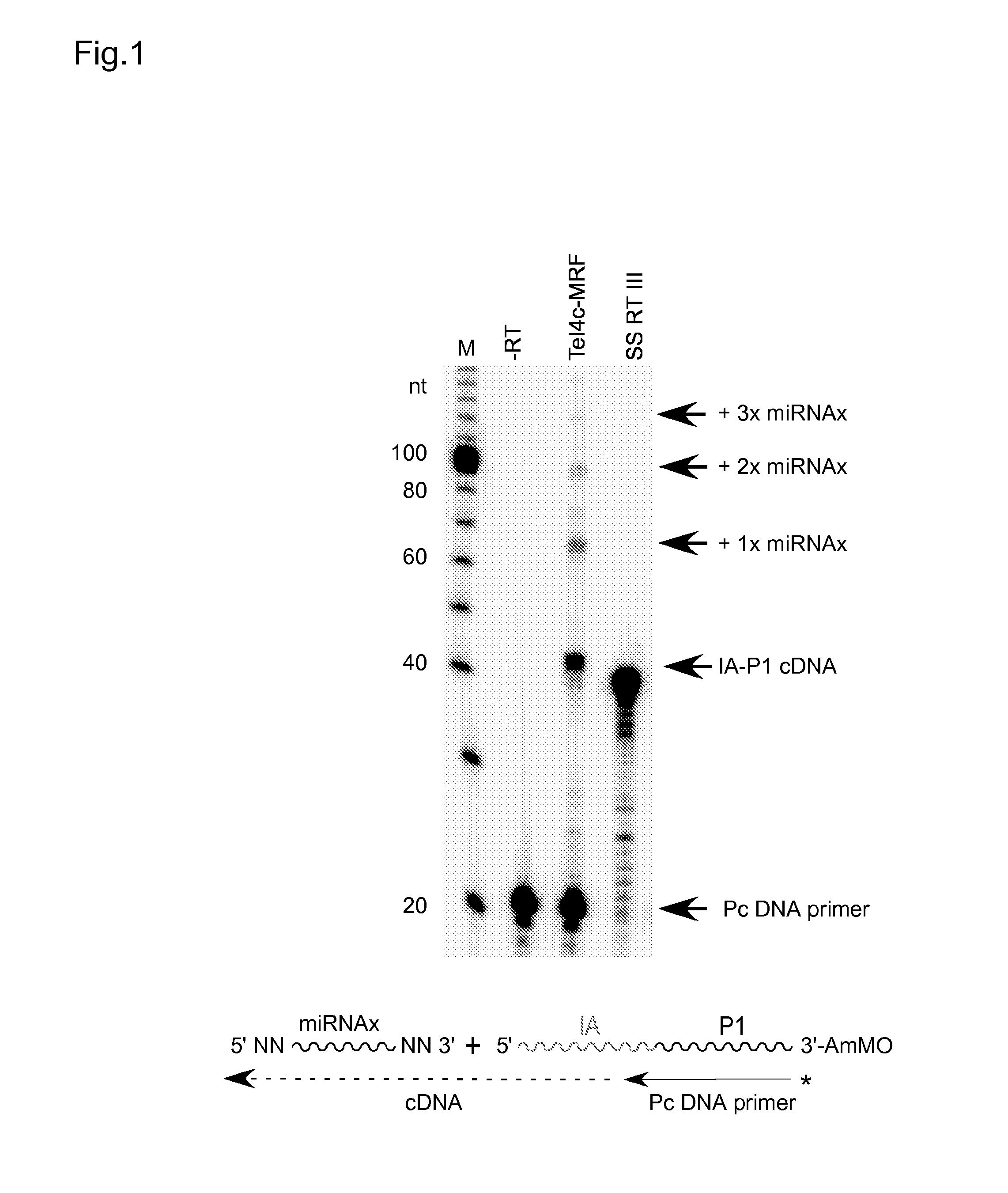
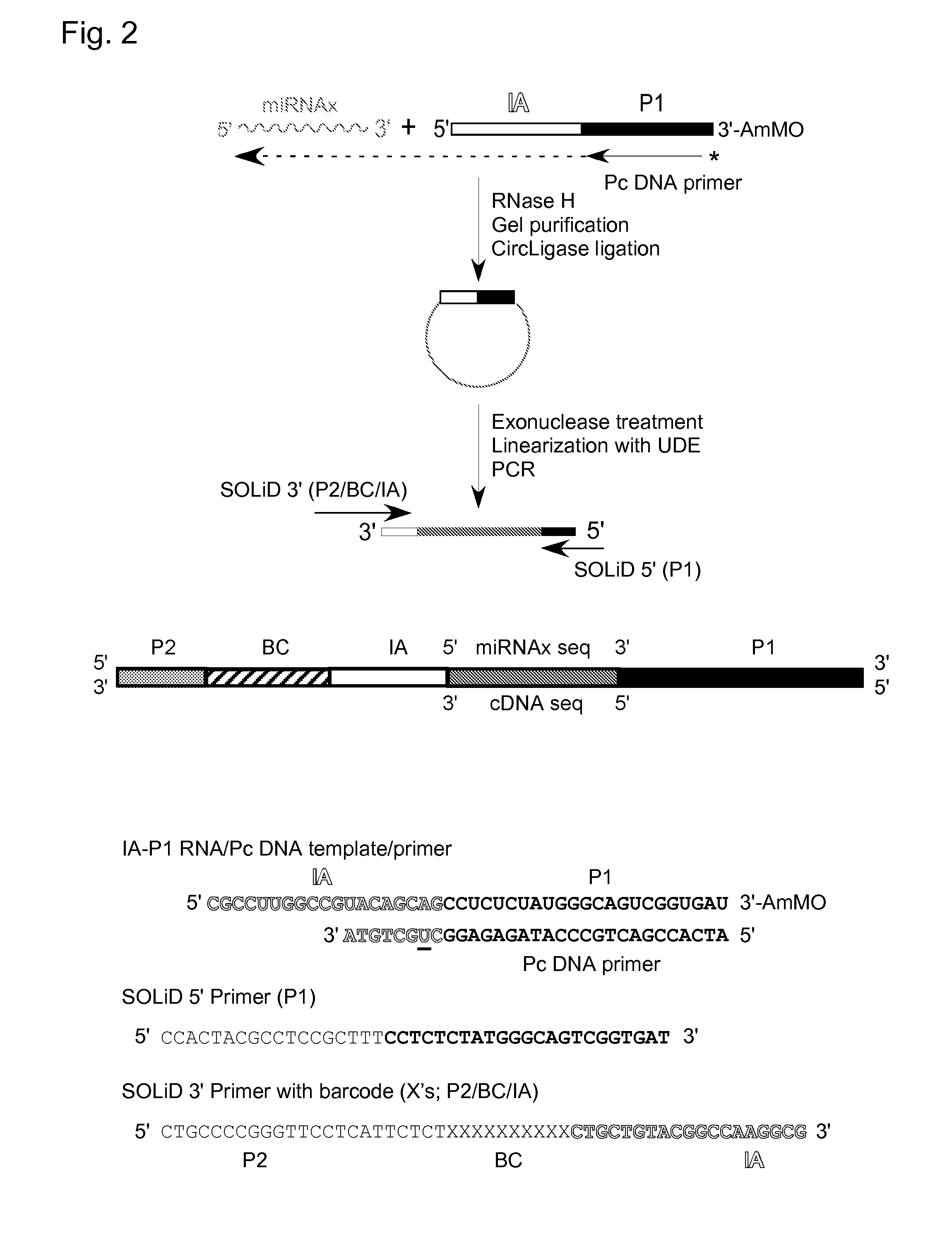
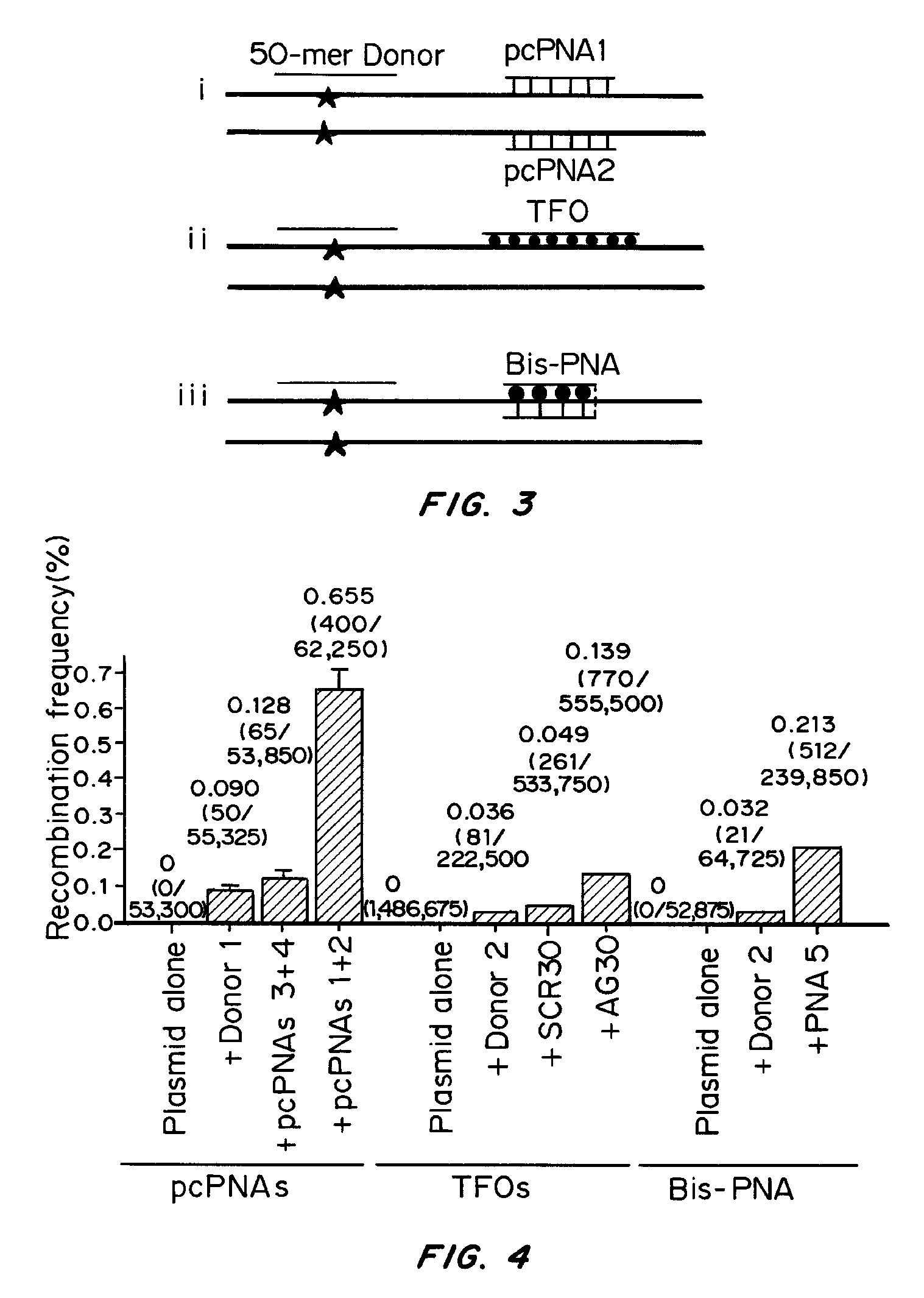
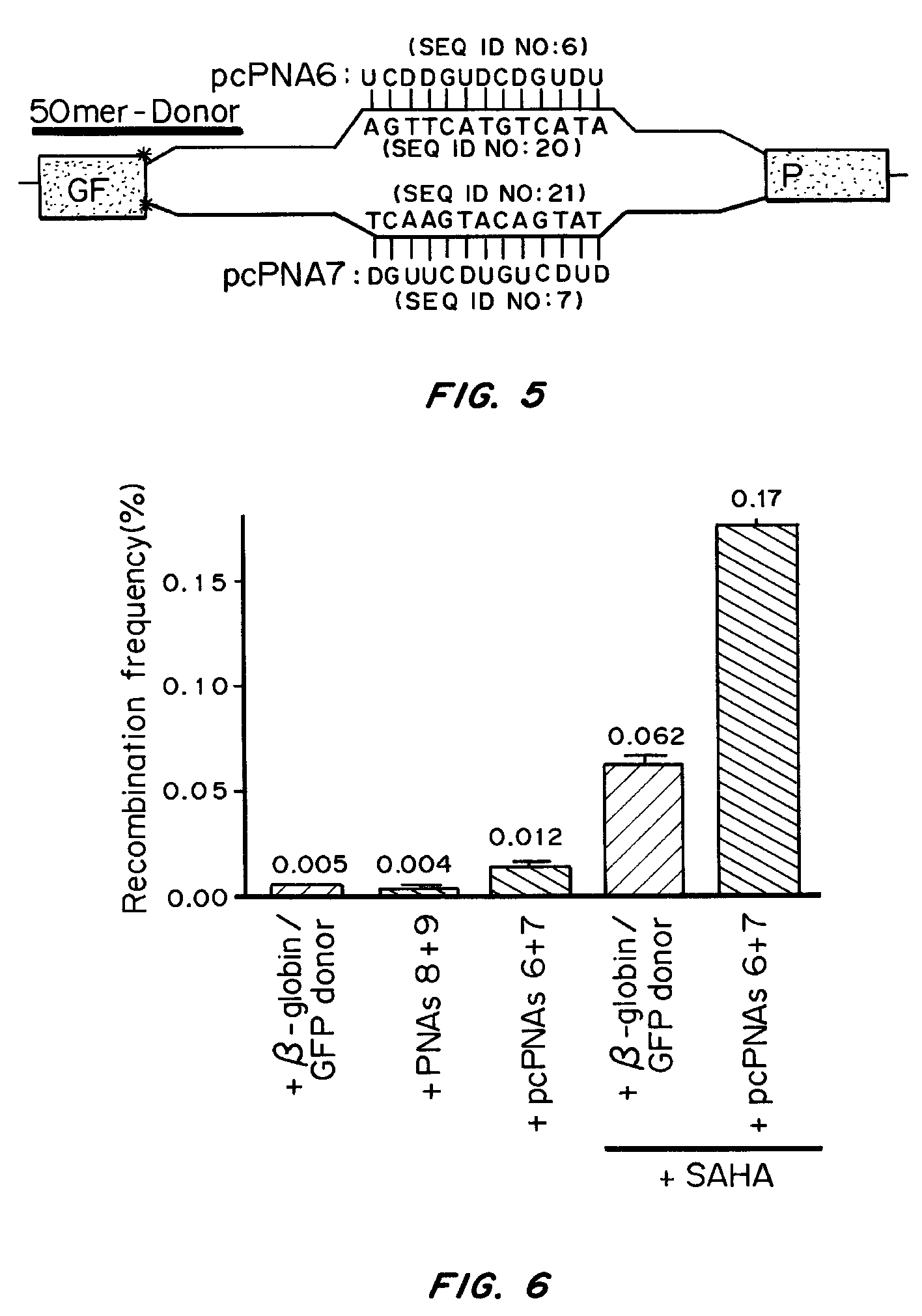
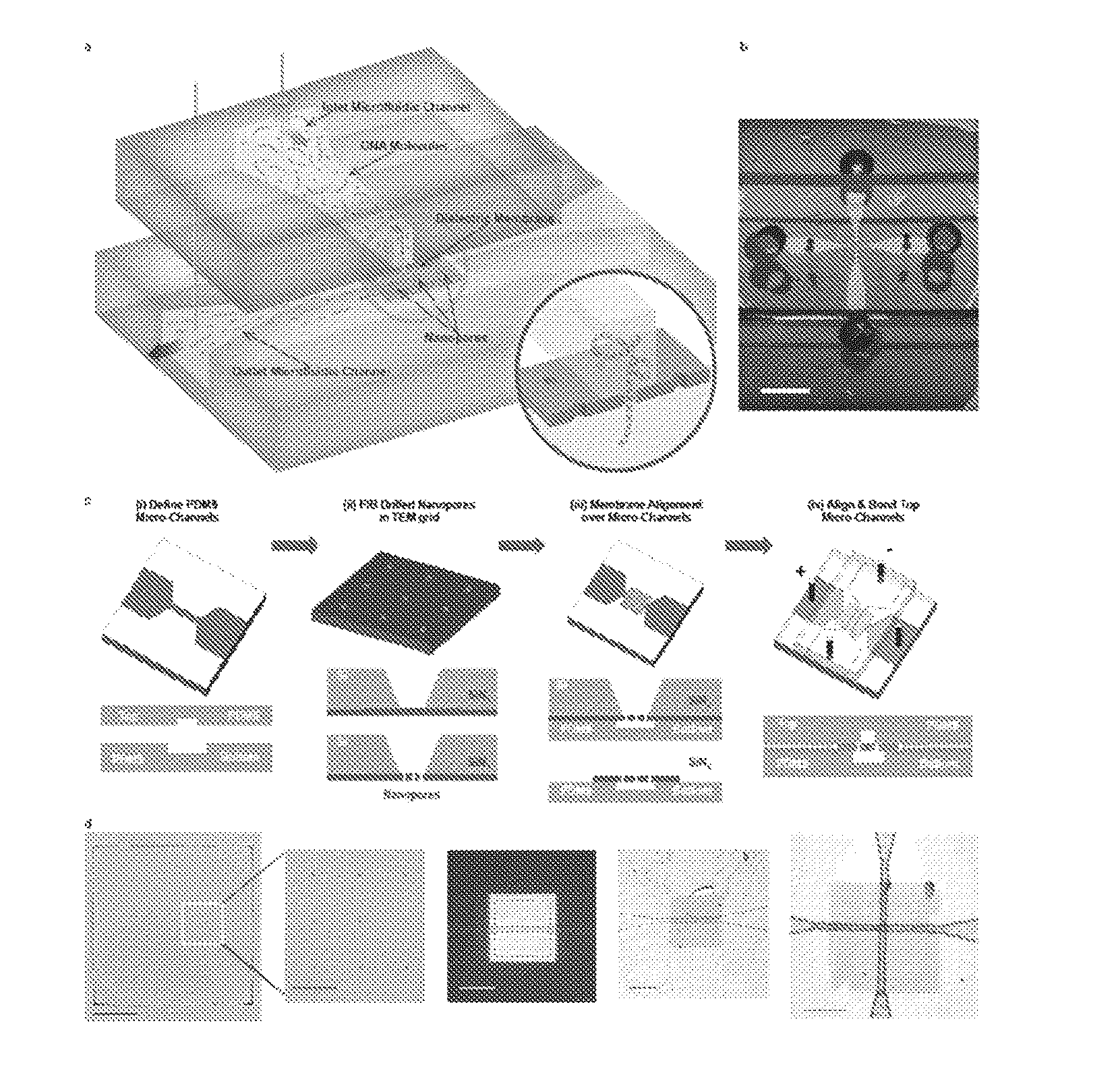
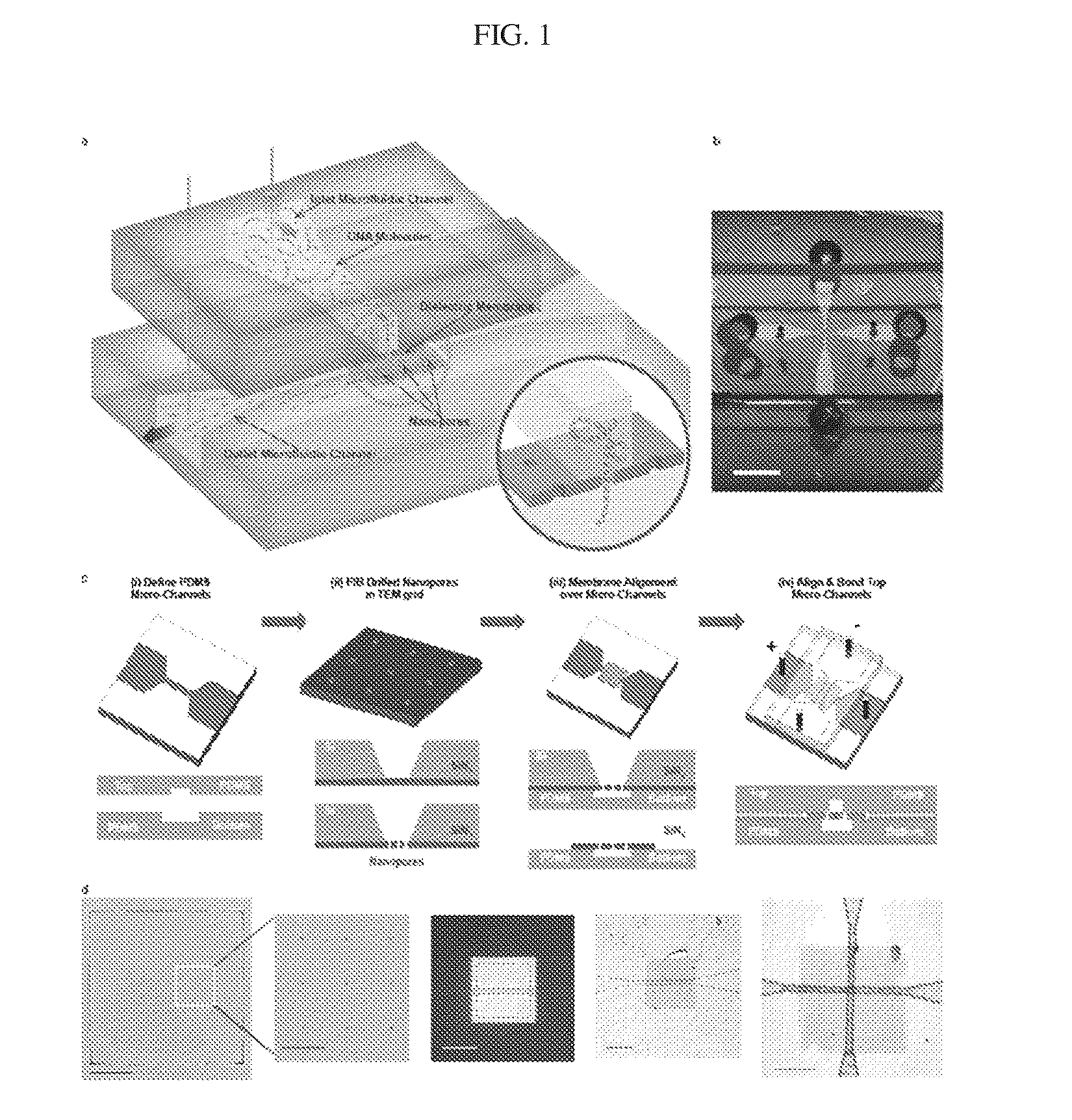
Patents
Literature
343 results about "DNA synthesis" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
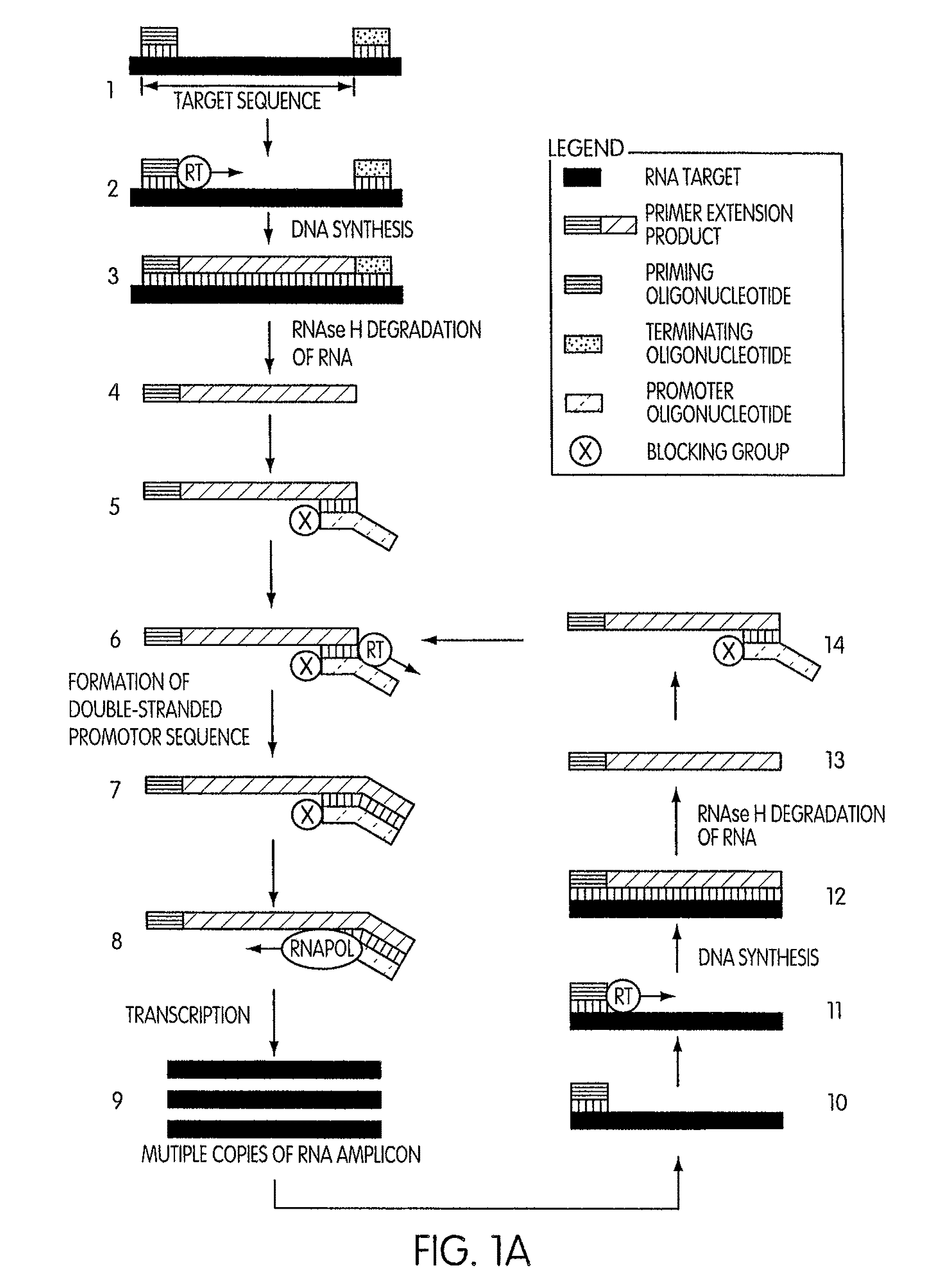
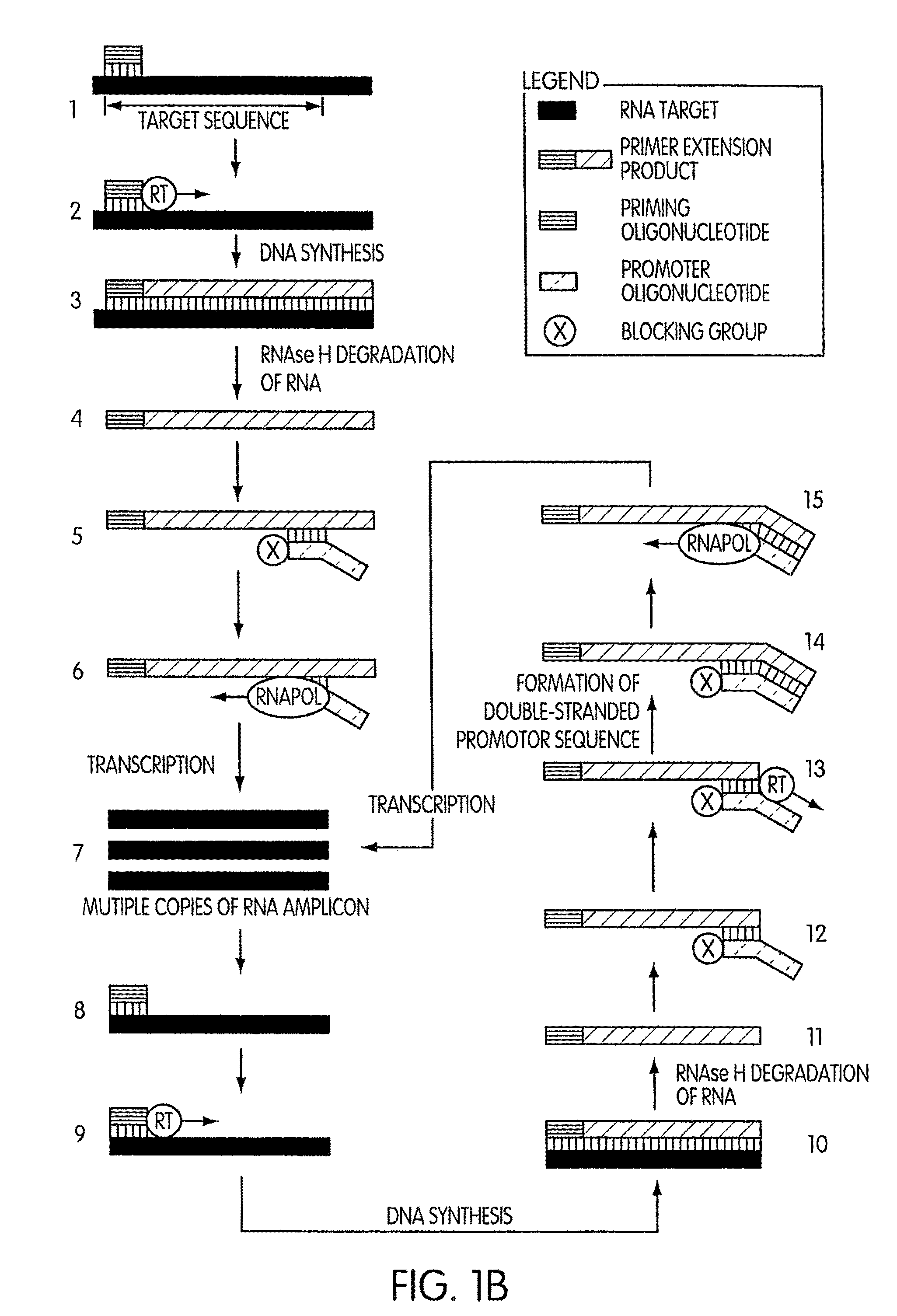
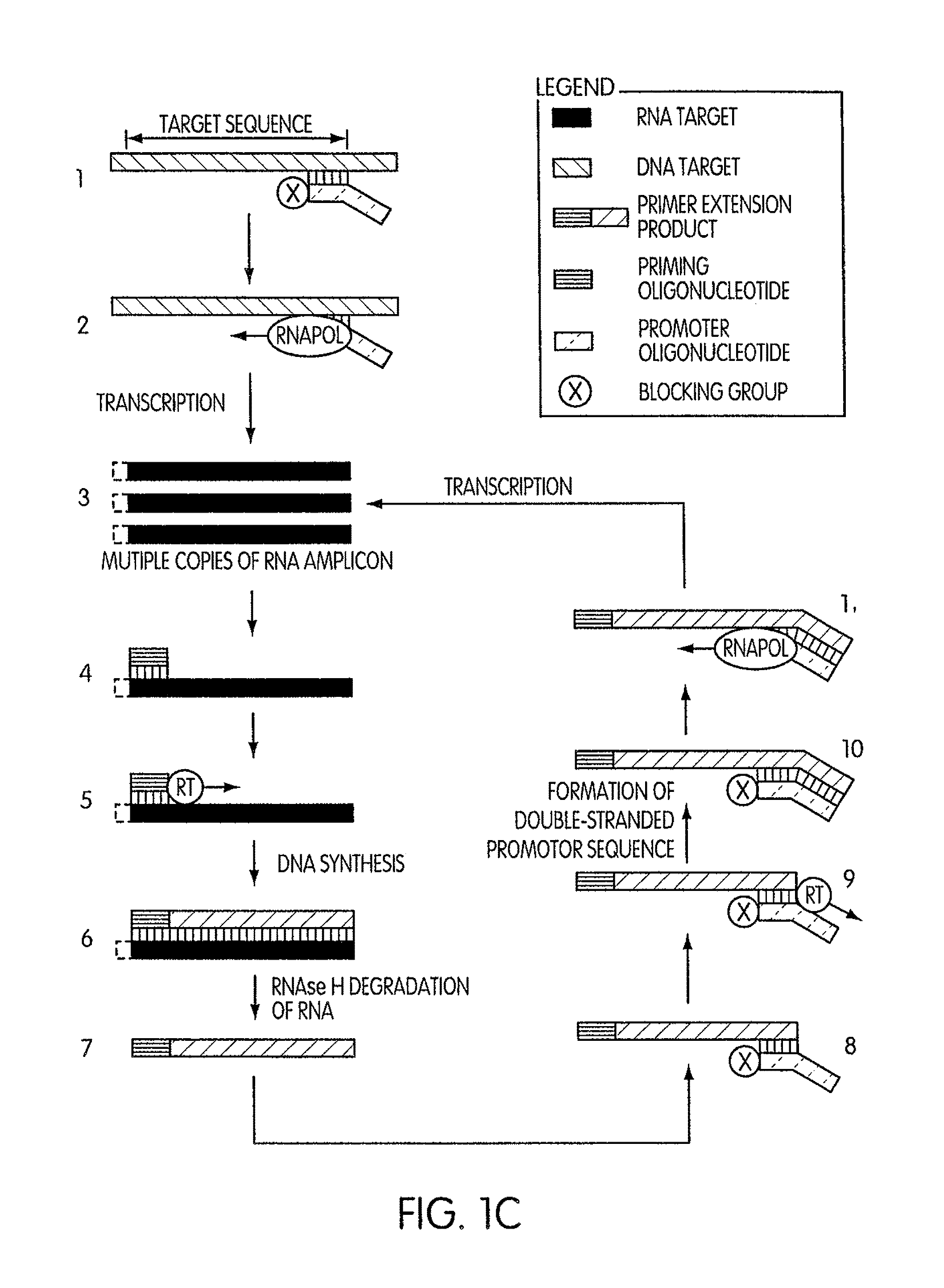
DNA synthesis is the natural or artificial creation of deoxyribonucleic acid (DNA) molecules. The term DNA synthesis can refer to DNA replication - DNA biosynthesis (in vivo DNA amplification), polymerase chain reaction - enzymatic DNA synthesis (in vitro DNA amplification) or gene synthesis - physically creating artificial gene sequences.
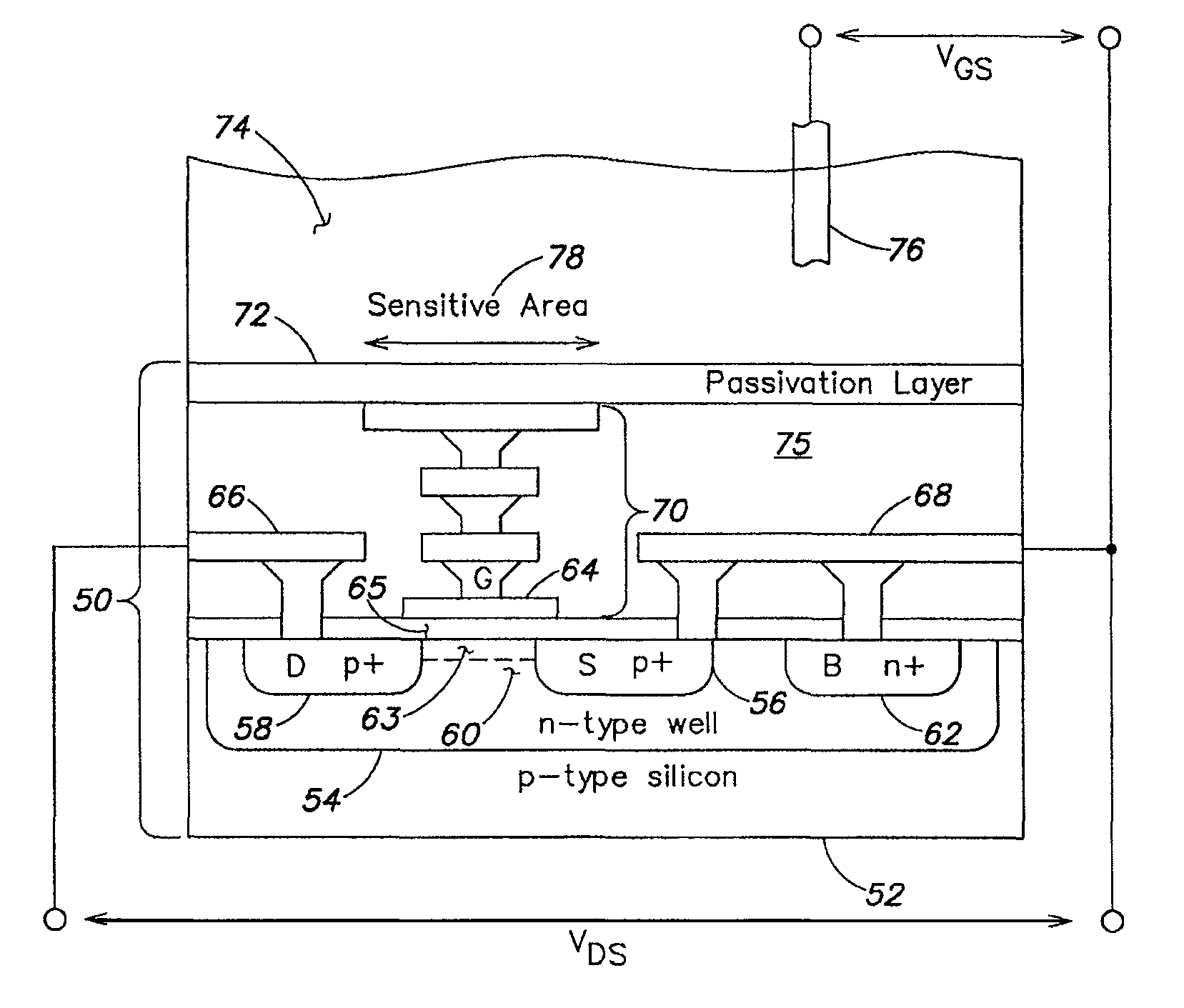
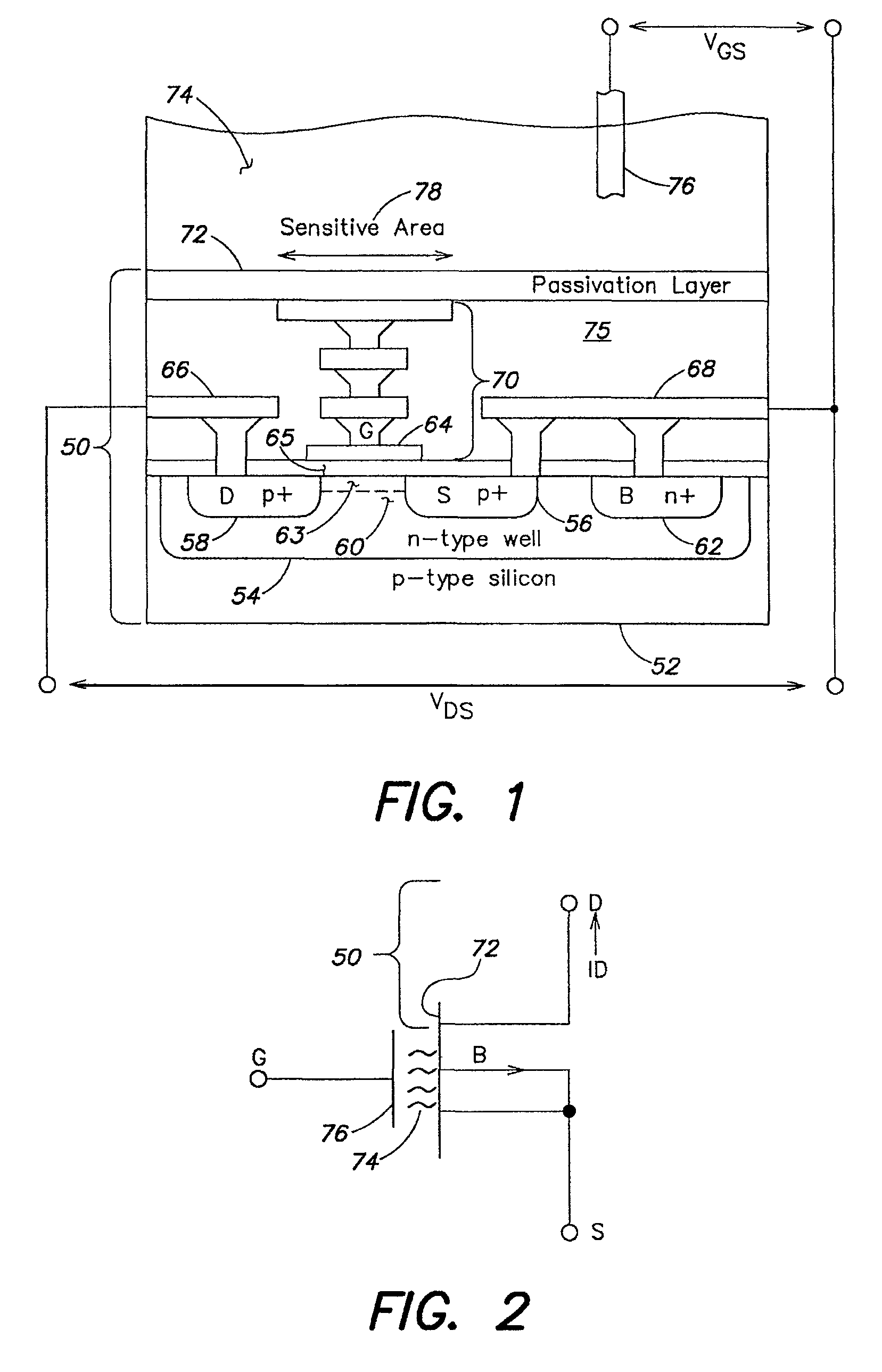
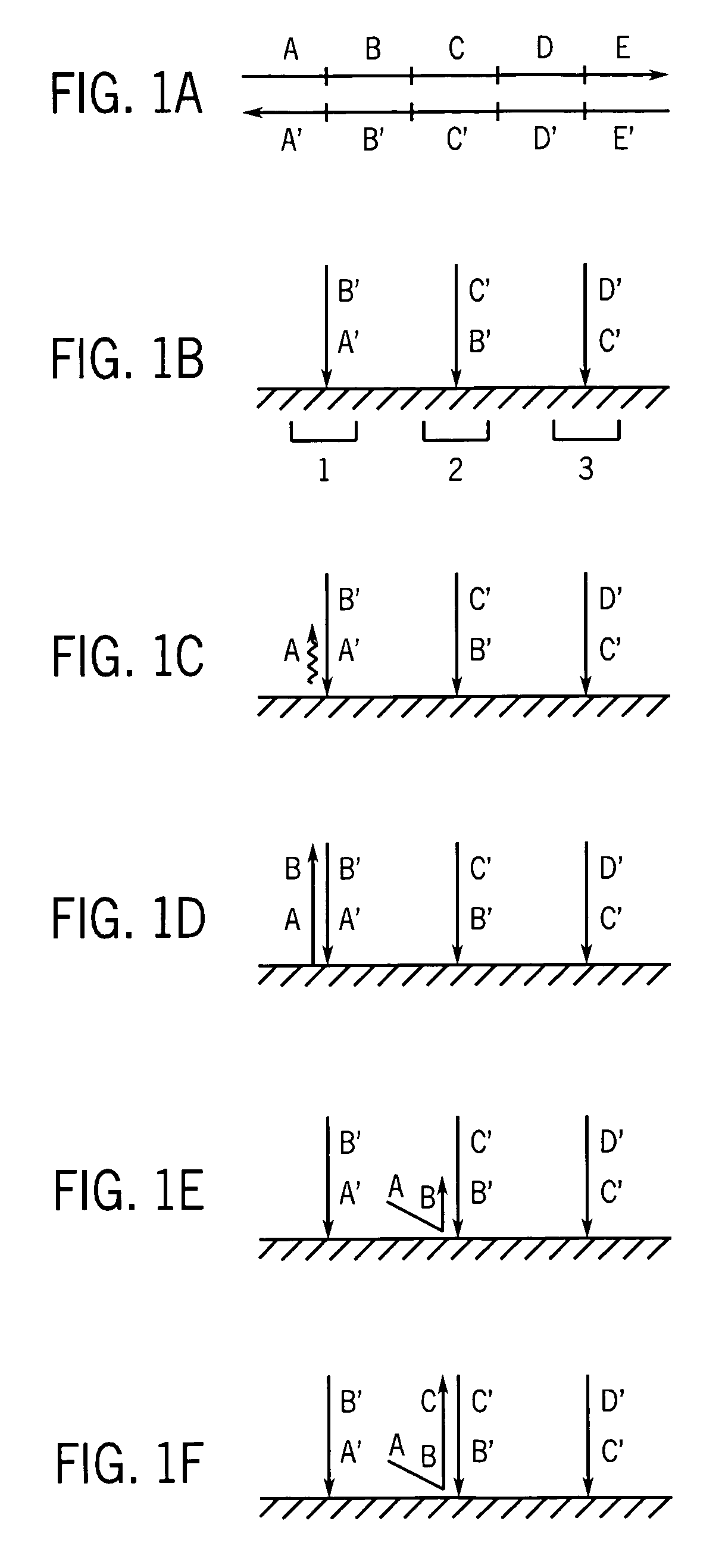
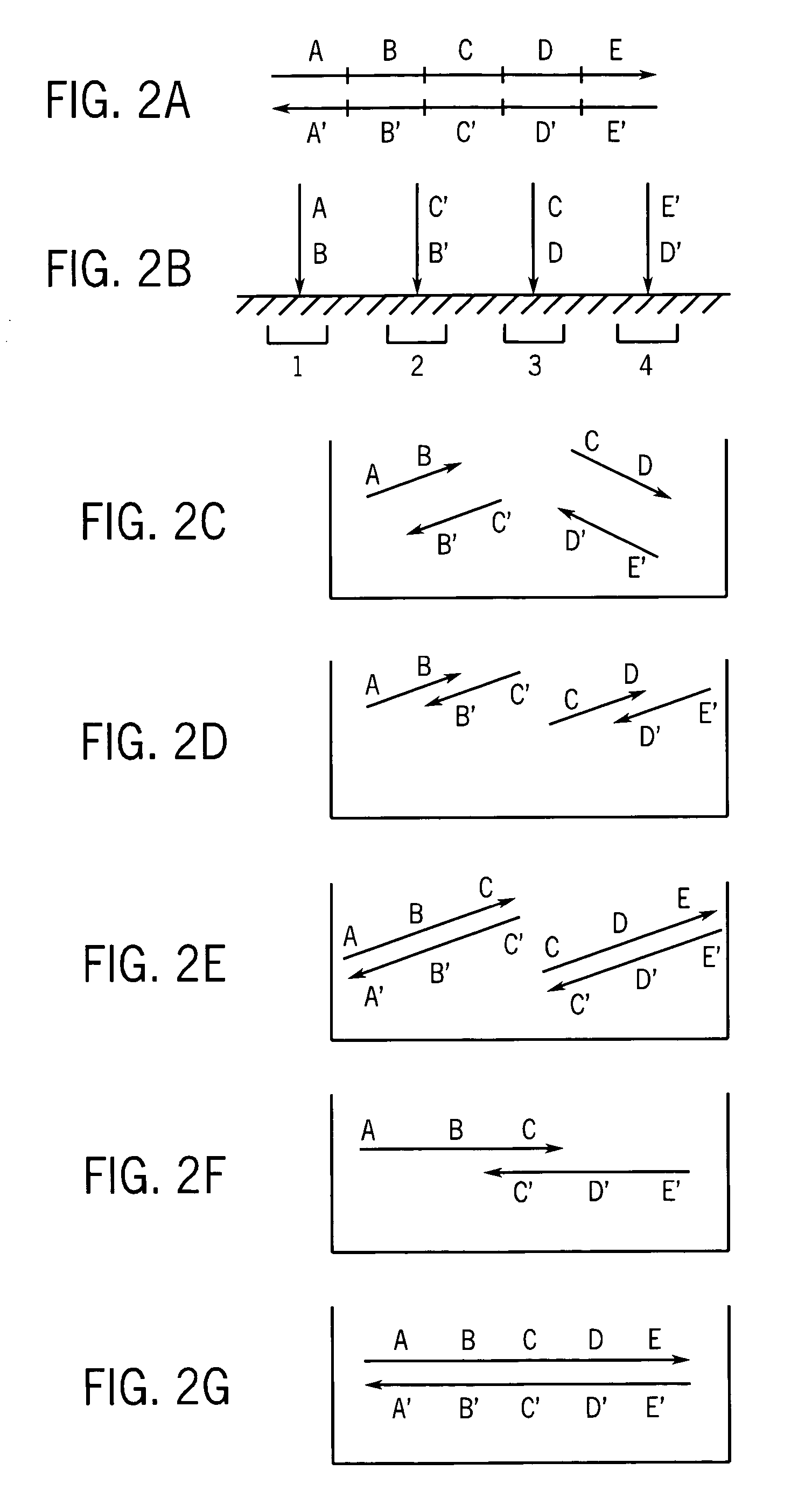
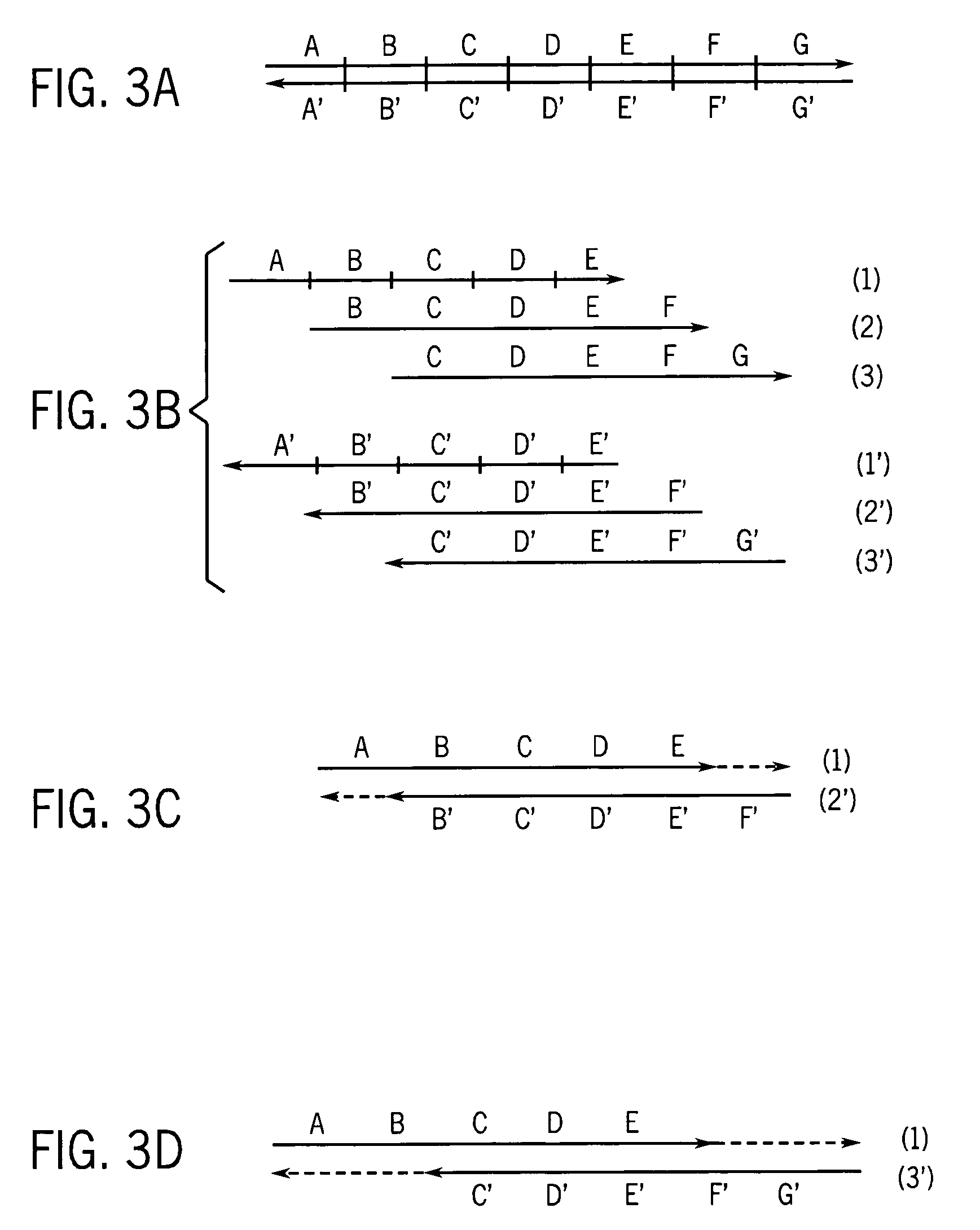
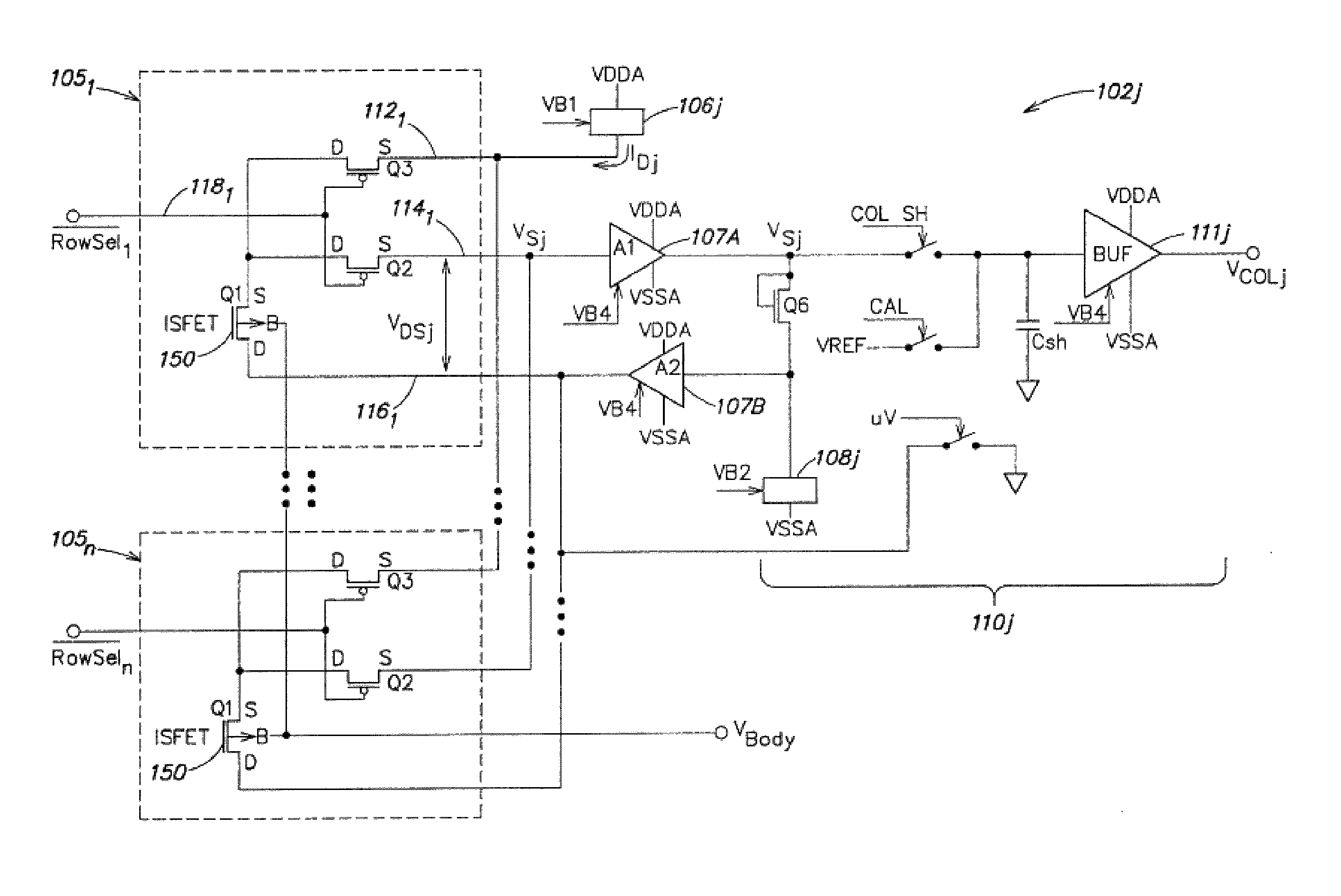
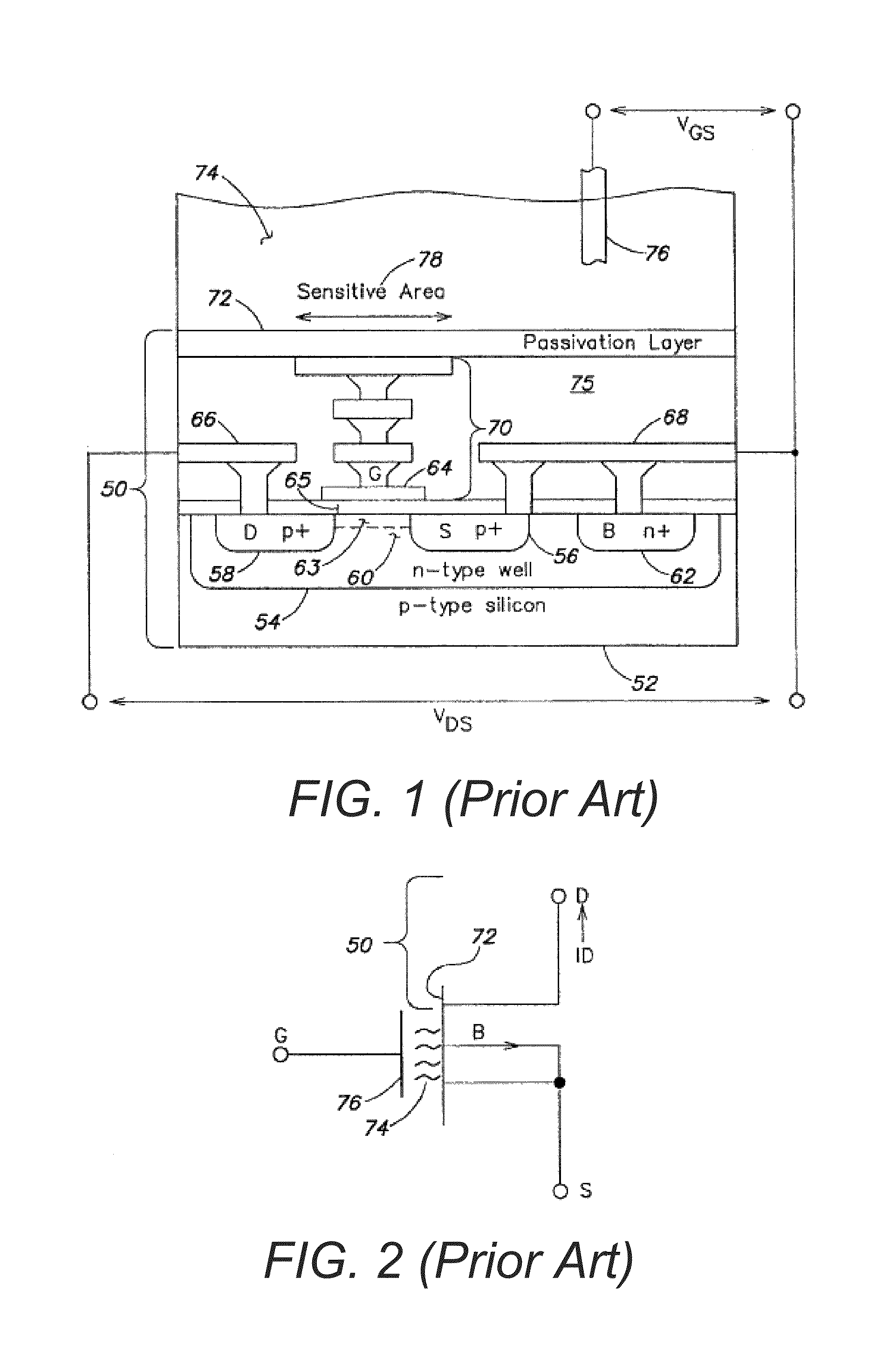
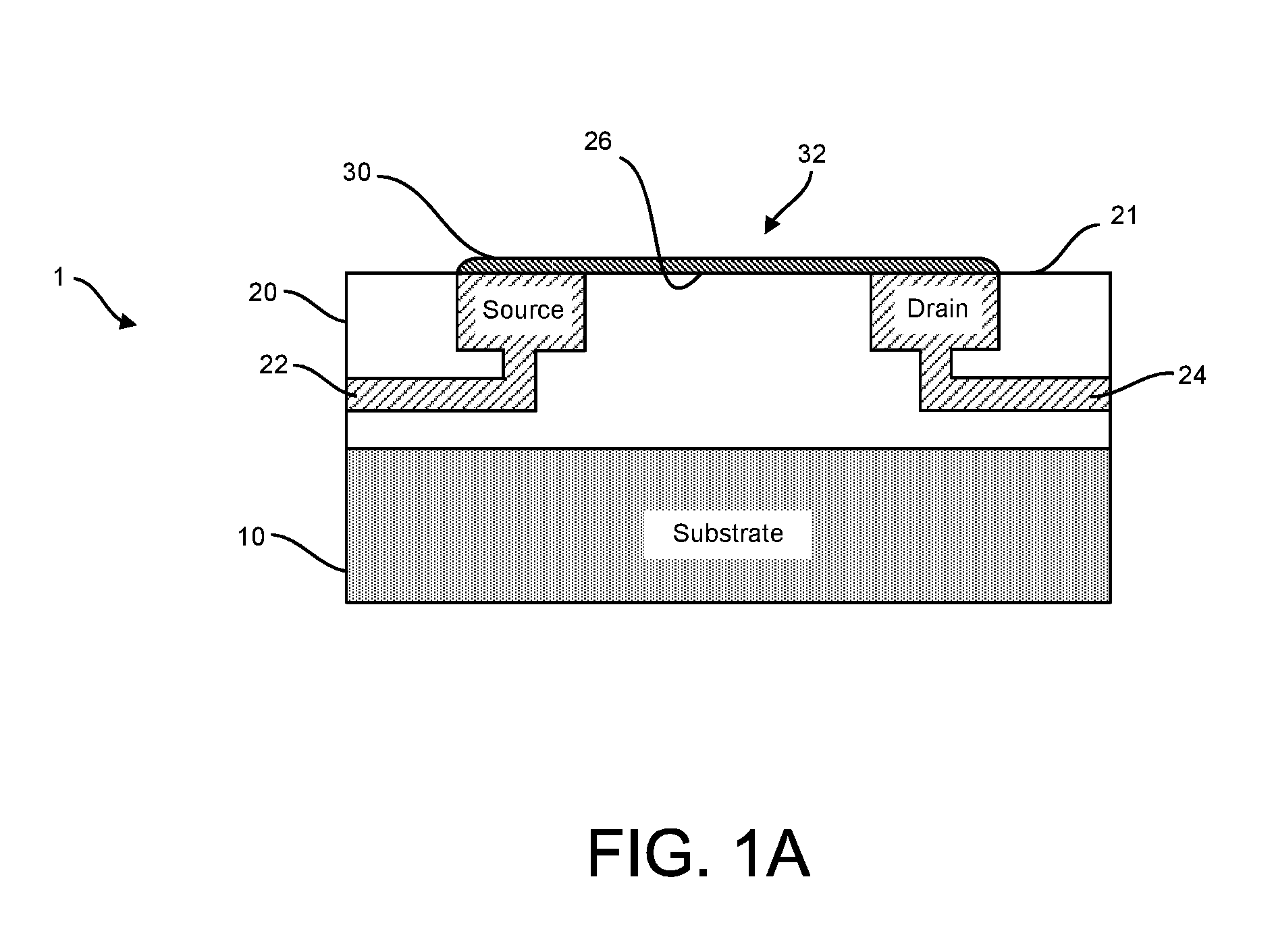
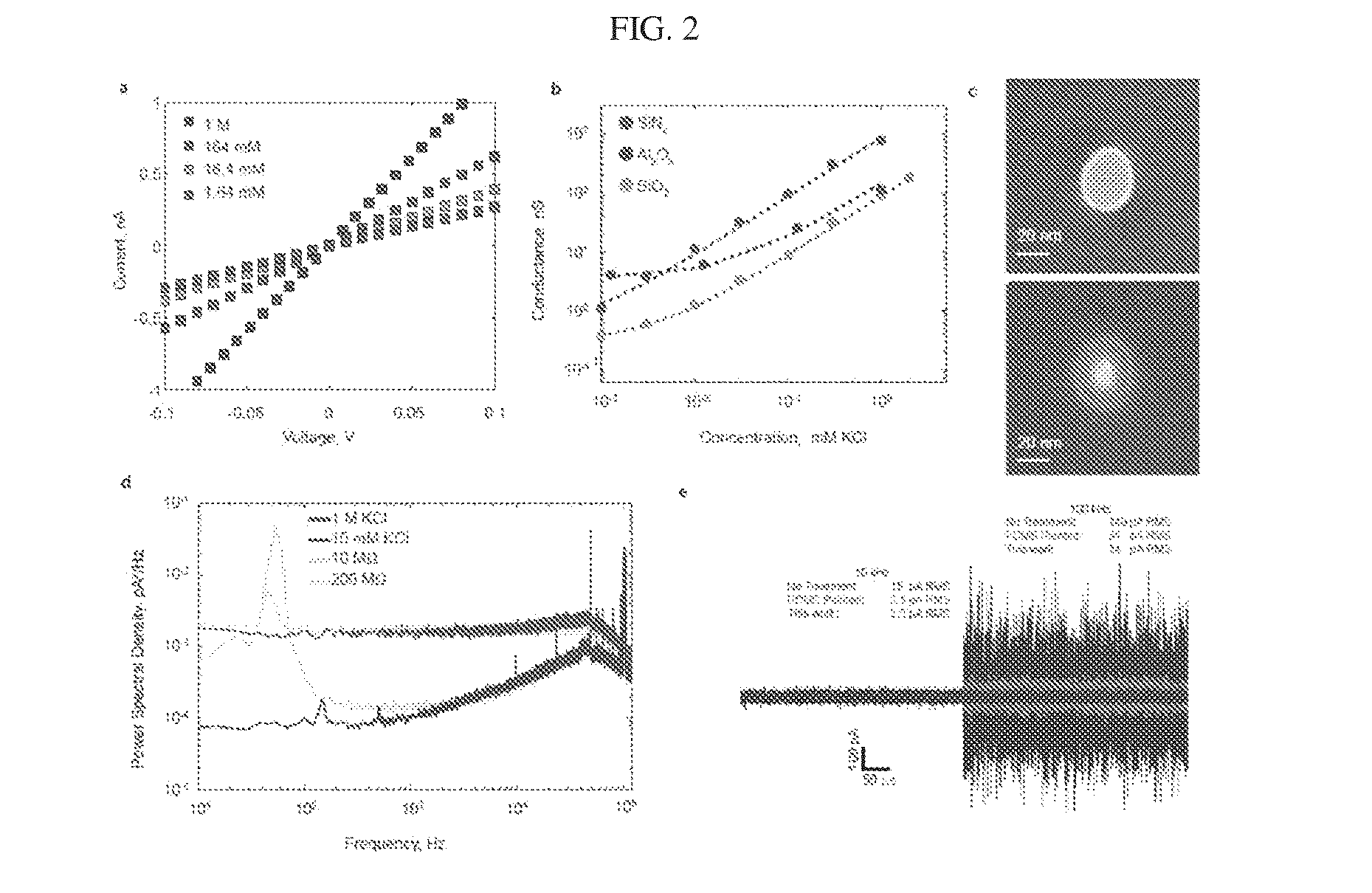
Methods and apparatus for measuring analytes using large scale FET arrays
ActiveUS7948015B2Reduce porosityHigh densityTransistorMicrobiological testing/measurementCMOSOrganismal Process
Methods and apparatus relating to very large scale FET arrays for analyte measurements. ChemFET (e.g., ISFET) arrays may be fabricated using conventional CMOS processing techniques based on improved FET pixel and array designs that increase measurement sensitivity and accuracy, and at the same time facilitate significantly small pixel sizes and dense arrays. Improved array control techniques provide for rapid data acquisition from large and dense arrays. Such arrays may be employed to detect a presence and / or concentration changes of various analyte types in a wide variety of chemical and / or biological processes. In one example, chemFET arrays facilitate DNA sequencing techniques based on monitoring changes in hydrogen ion concentration (pH), changes in other analyte concentration, and / or binding events associated with chemical processes relating to DNA synthesis.
Owner:LIFE TECH CORP
Methods for high fidelity production of long nucleic acid molecules with error control
InactiveUS20050227235A1Optimization orderIncrease opportunitiesSequential/parallel process reactionsSugar derivativesUser controlDNA synthesis
This invention generally relates to nucleic acid synthesis, in particular DNA synthesis. More particularly, the invention relates to the production of long nucleic acid molecules with precise user control over sequence content. This invention also relates to the prevention and / or removal of errors within nucleic acid molecules.
Owner:MASSACHUSETTS INST OF TECH
Compositions for enhancing DNA synthesis, DNA polymerase-related factors and utilization thereof
ActiveUS7384739B2High synthetic activityEnhancing PCR success rateMicrobiological testing/measurementTransferasesEnzymatic synthesisPHA polymerase
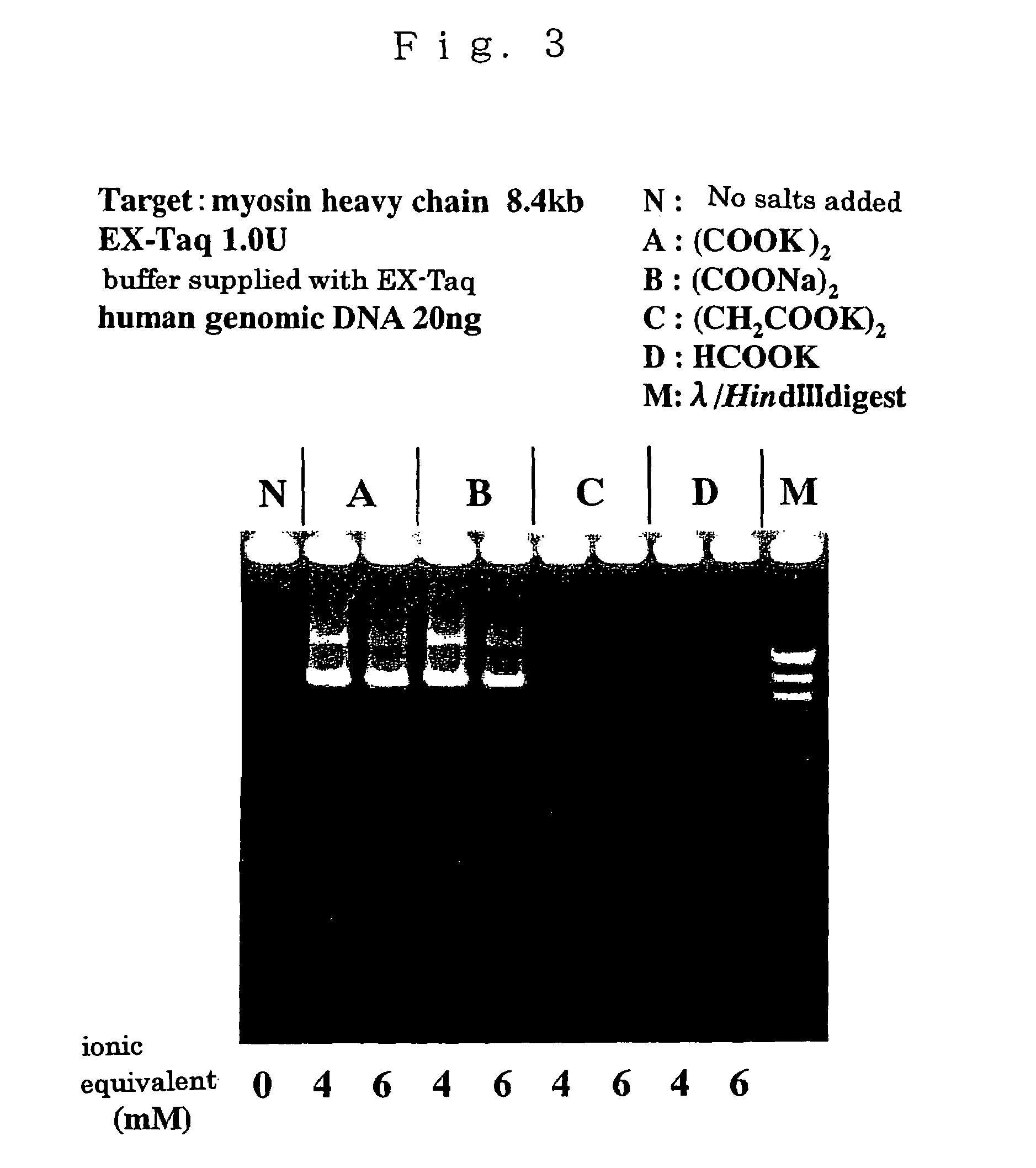
The invention provides methods, kits, and compositions for enhancing synthesis of DNA involving a carboxylate ion-supplying substance that is effective in promoting DNA synthesis in enzymatic DNA synthesis reactions. The invention further provides a thermostable DNA polymerase-related factor derived from Thermococcus species, which has an activity to promote the DNA synthesis activity of DNA polymerase or which binds to DNA polymerase.
Owner:TOYOBO CO LTD
Methods for high fidelity production of long nucleic acid molecules with error control
InactiveUS7932025B2Increase opportunitiesEasy to joinSequential/parallel process reactionsSugar derivativesUser controlDNA synthesis
This invention generally relates to nucleic acid synthesis, in particular DNA synthesis. More particularly, the invention relates to the production of long nucleic acid molecules with precise user control over sequence content. This invention also relates to the prevention and / or removal of errors within nucleic acid molecules.
Owner:MASSACHUSETTS INST OF TECH
Methods for high fidelity production of long nucleic acid molecules
InactiveUS7879580B2Increase opportunitiesEasy to joinSugar derivativesMicrobiological testing/measurementUser controlDNA synthesis
This invention generally relates to nucleic acid synthesis, in particular DNA synthesis. More particularly, the invention relates to the production of long nucleic acid molecules with precise user control over sequence content. This invention also relates to the prevention and / or removal of errors within nucleic acid molecules.
Owner:MASSACHUSETTS INST OF TECH
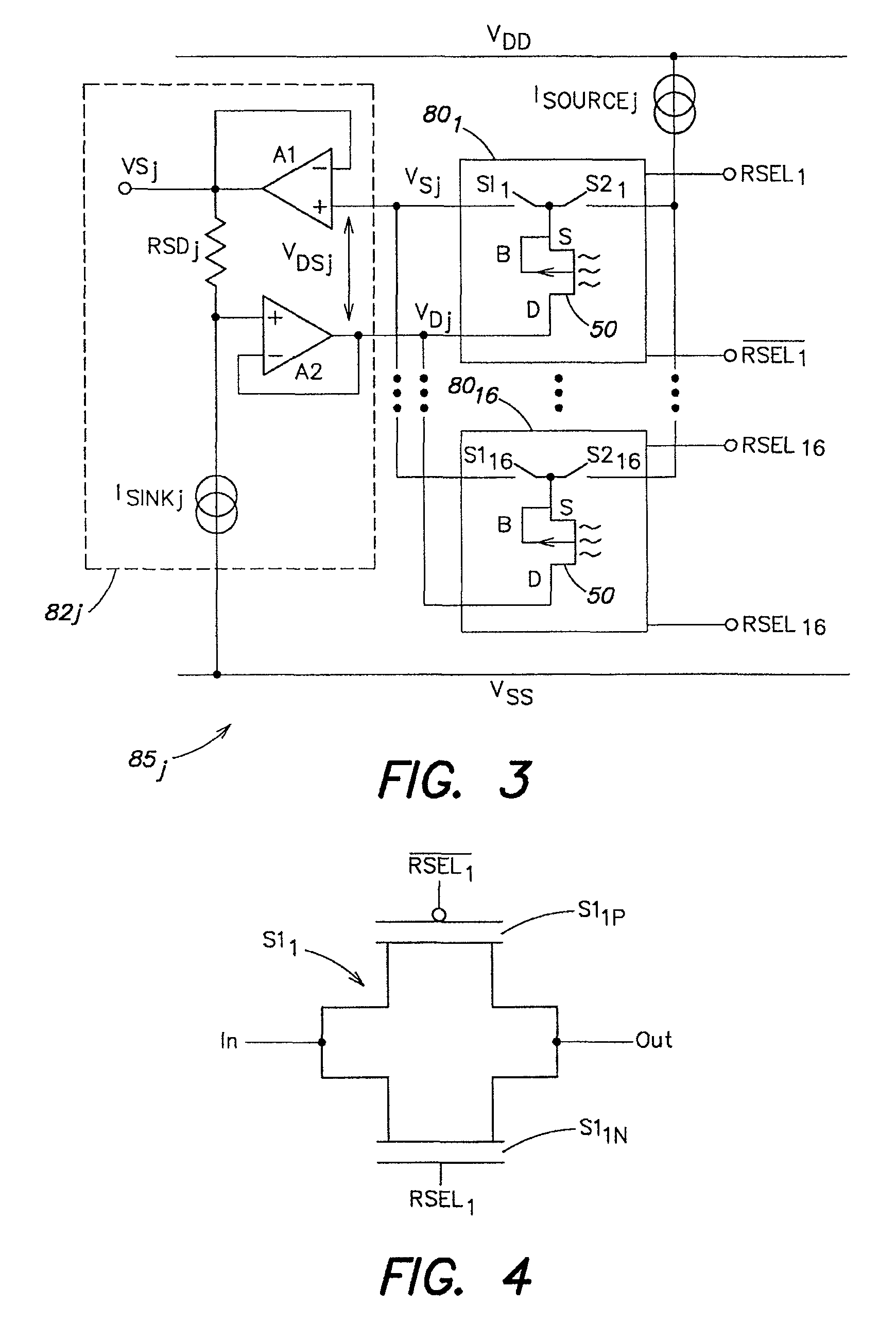
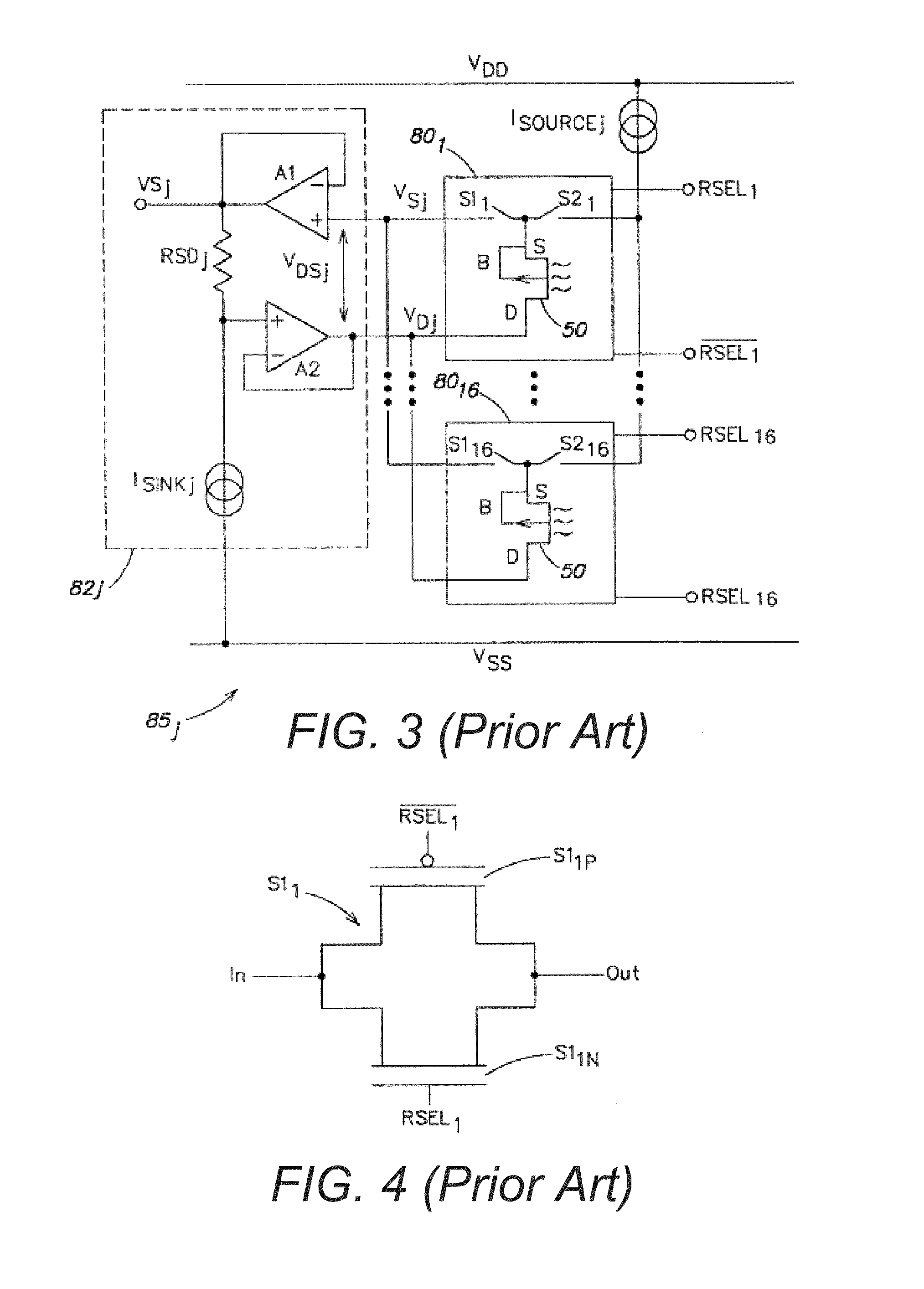
Methods for operating chemically-sensitive sample and hold sensors
ActiveUS8496802B2Facilitate rapid acquisition of dataSmall sizeTransistorWeather/light/corrosion resistanceCMOSAnalyte
Methods and apparatus relating to very large scale FET arrays for analyte measurements. ChemFET (e.g., ISFET) arrays may be fabricated using conventional CMOS processing techniques based on improved FET pixel and array designs that increase measurement sensitivity and accuracy, and at the same time facilitate significantly small pixel sizes and dense arrays. Improved array control techniques provide for rapid data acquisition from large and dense arrays. Such arrays may be employed to detect a presence and / or concentration changes of various analyte types in a wide variety of chemical and / or biological processes. In one example, chemFET arrays facilitate DNA sequencing techniques based on monitoring changes in hydrogen ion concentration (pH), changes in other analyte concentration, and / or binding events associated with chemical processes relating to DNA synthesis.
Owner:LIFE TECH CORP
Method for amplifying nucleic acid sequence
A convenient and effective method for amplifying a nucleic acid sequence characterized by effecting a DNA synthesis reaction in the presence of chimeric oligonucleotide primers; a method for supplying a large amount of DNA amplification fragments; an effective method for amplifying a nucleic acid sequence by combining the above method with another nucleic acid sequence amplification method; a method for detecting a nucleic acid sequence for detecting or quantitating a microorganism such as a virus, a bacterium, a fungus or a yeast; and a method for detecting a DNA amplification fragment obtained by the above method in situ.
Owner:TAKARA HOLDINGS
Method to clone mRNAs
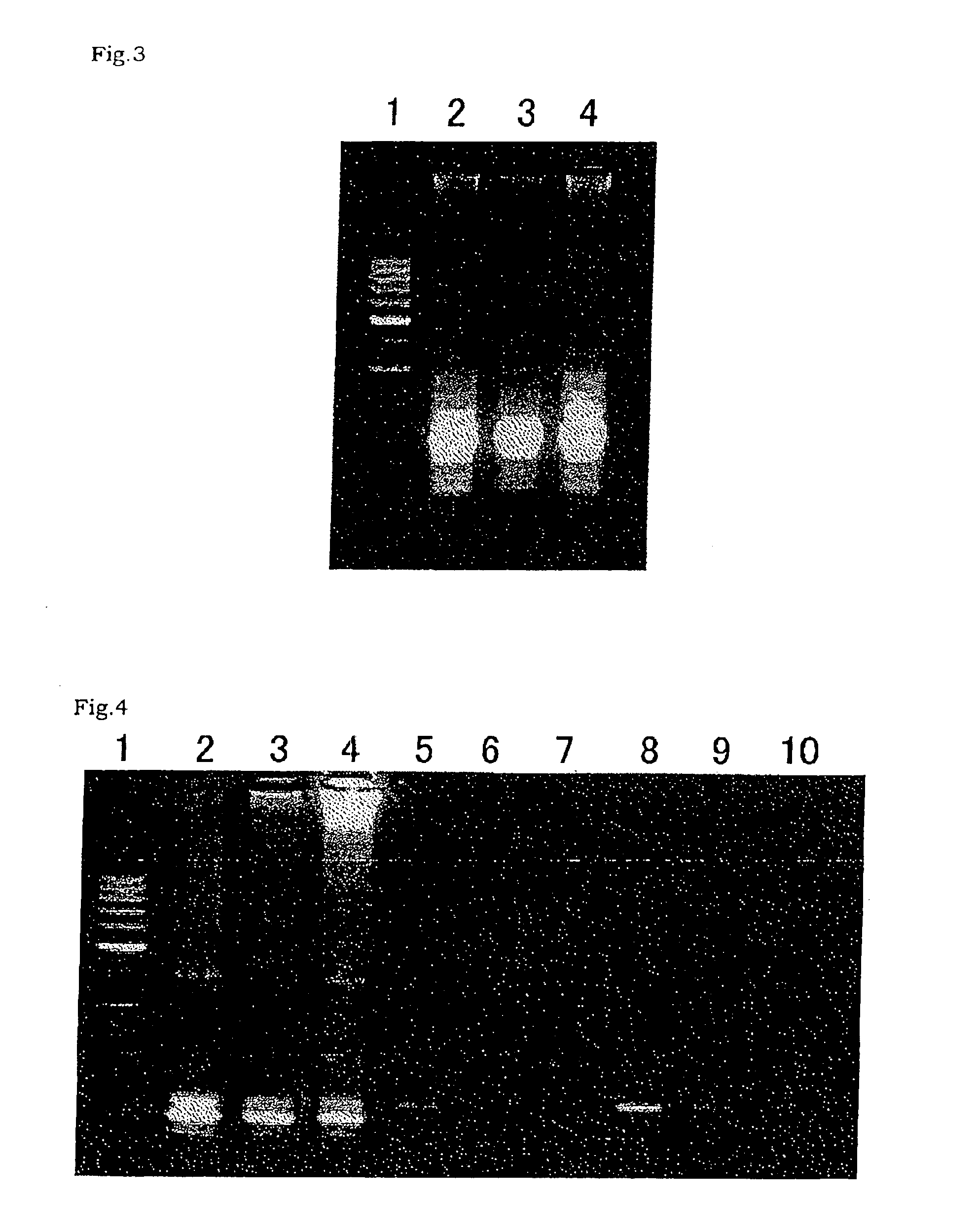
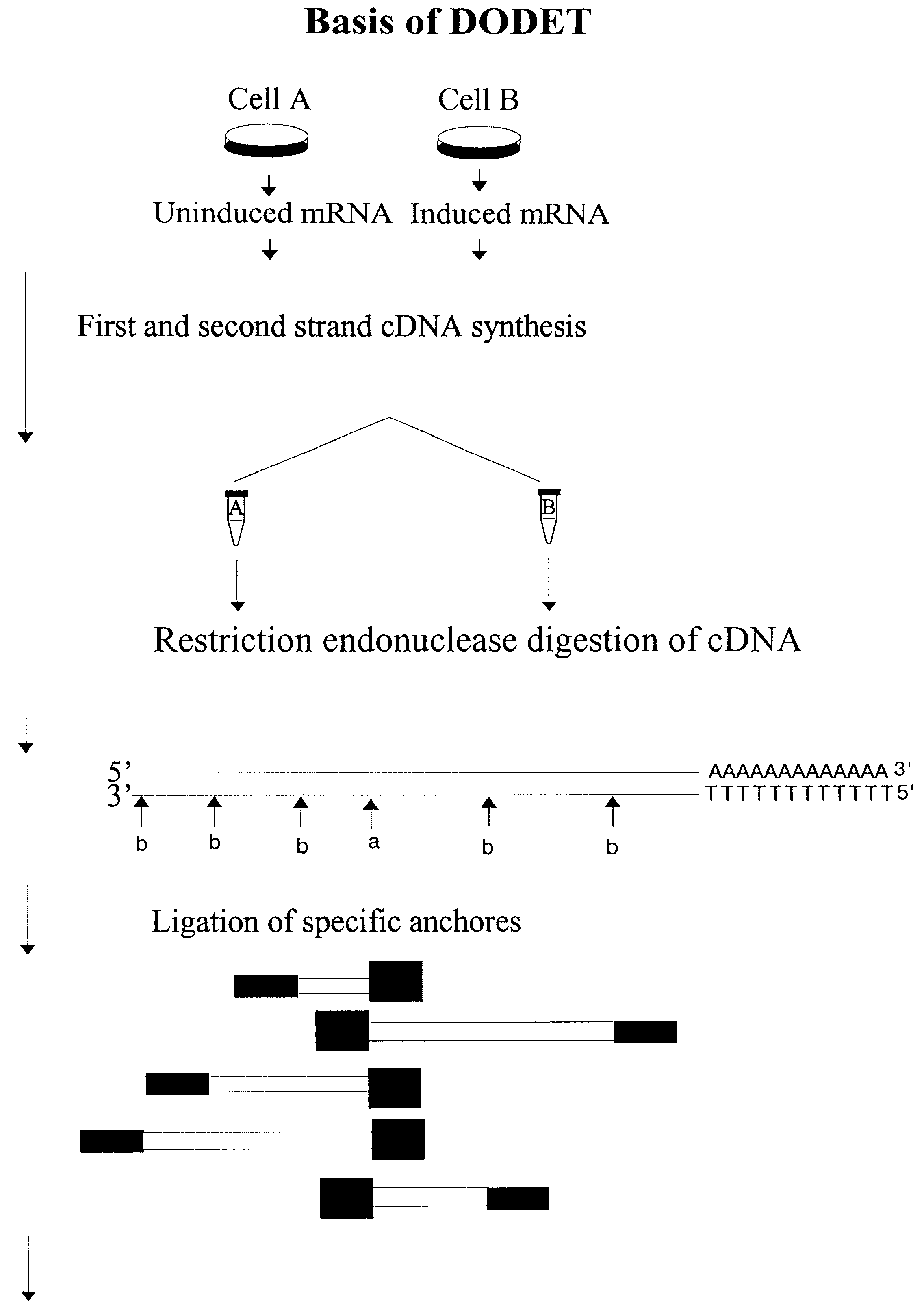
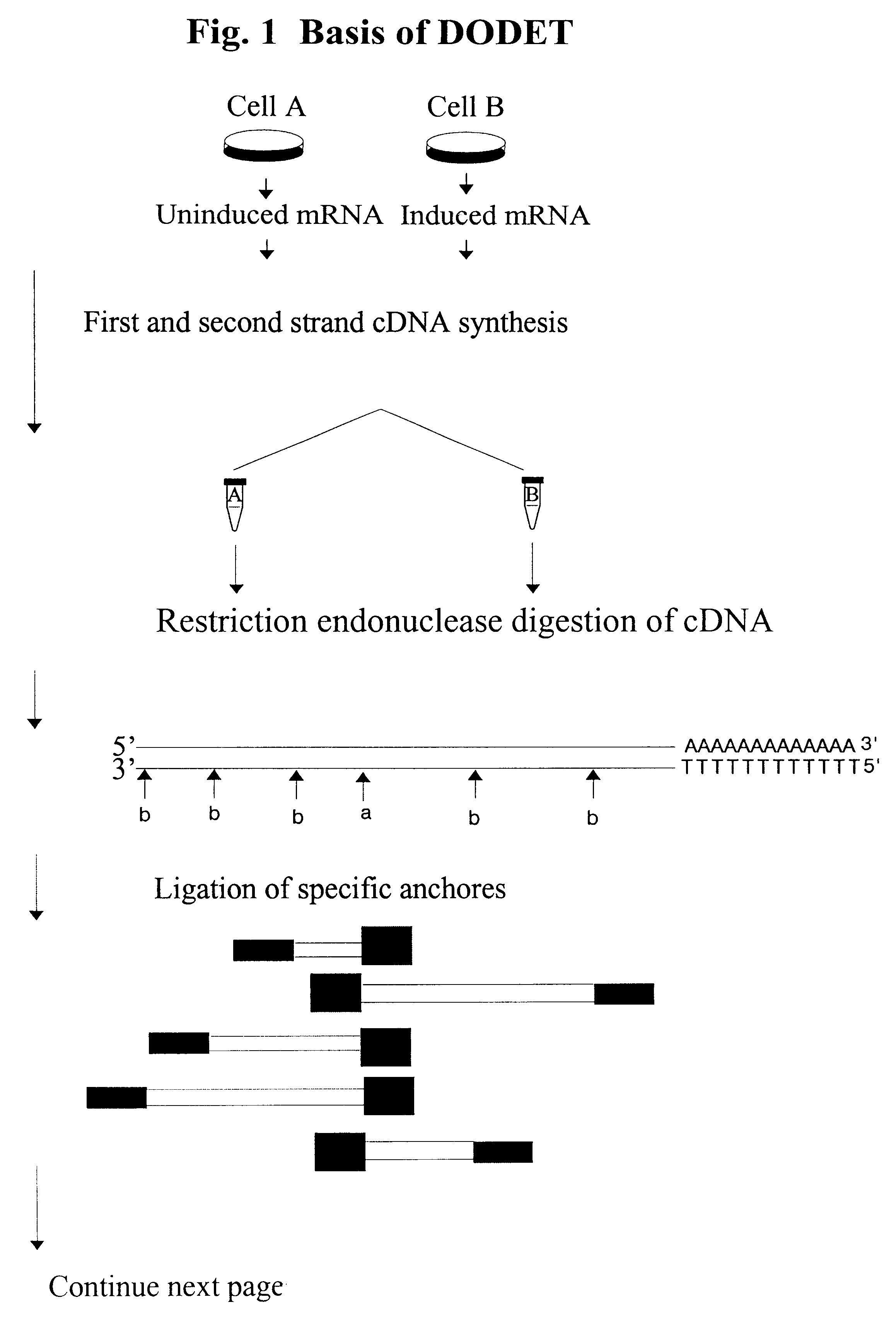
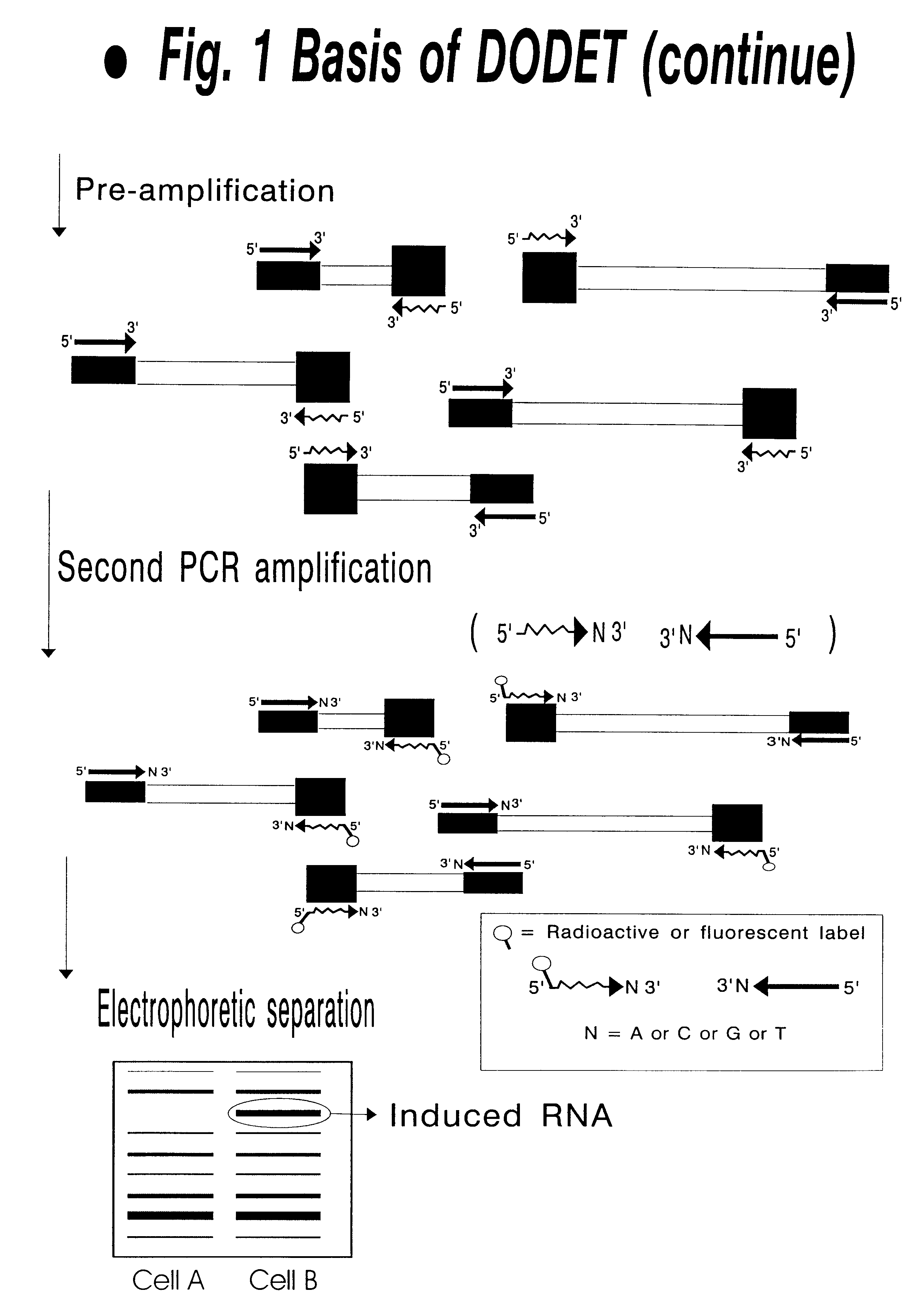
InactiveUS6261770B1Convenient verificationHigh homologySugar derivativesMicrobiological testing/measurementNucleotideNucleotide sequencing
Disclosed and claimed is a method for preparing a normalized sub-divided library of amplified cDNA fragments from the coding region of mRNAs contained in a sample. The method includes the steps of: a) subjecting the mRNA population to reverse transcription using at least one cDNA primer, thereby obtaining first strand cDNA fragments, b) synthesizing second strand cDNA complementary to the first strand cDNA fragments by use of the first strand DNA fragments as templates, thereby obtaining double stranded cDNA fragments, c) digesting the double stranded cDNA fragments with at least one restriction endonuclease, the endonuclease leaving protruding sticky ends of similar size at the termini of the DNA after digestion, thereby obtaining cleaved cDNA fragments, d) adding at least two adapter fragments containing known sequences to the cleaved cDNA fragments obtained in step c), the at least two adapter fragments being able to bind specifically to the sticky ends of the double stranded cDNA produced in step c), the one adapter fragment being able to anneal to the primer having formula I in step f), the second adapter fragment being a termination fragment introducing a block against DNA polymerization in the 5'->3' direction setting out from said termination fragment and the termination fragment being unable to anneal to any primer of the at least two primer sets in step f) during the molecular amplification procedure, the at least two adapter fragments being ligated to the cleaved cDNA fragments obtained in step c) so as to obtain ligated cDNA fragments, e) sub-dividing the ligated cDNA fragments obtained in step d) into 4n1 pools where 1<=n1<=4, and f) subjecting each pool of ligated cDNA fragments obtained in step e) to a molecular amplification procedure so as to obtain amplified cDNA fragments, wherein is used, for an adapter fragment used in step d), a set of amplification primers having the general formula Iwherein Com is a sequence complementary to at least the 5'-end of an adapter fragment which is ligated to the 3'-end of a cleaved cDNA fragment, N is A, G, T, or C, the one primer having the general formula I where n1=0, and the second primer having the general formula I where 1<=n1<=4, the second primer being capable of priming amplification of any nucleotide sequence ligated in its 3'-end to the adapter fragment complementary in its 5'-end to Com.
Owner:AZIGN BIOSCI
Dual Function Primers for Amplifying DNA and Methods of Use
InactiveUS20090068643A1Enabling detectionSugar derivativesMicrobiological testing/measurementFluorescenceAmplification dna
The present invention provides novel nucleotide compositions that enable the detection of DNA synthesis products and methods for use thereof. In one embodiment, the method can be used in PCR and allows the progress of the reaction to be monitored as it occurs. In one embodiment, the invention employs at least one fluorescence-quenched oligonucleotide that can prime DNA extension reactions. In a second embodiment, the invention employs at least one fluorescence-quenched oligonucleotide that can function as a template for DNA extension reactions. In both embodiments, the oligonucleotide also functions as a probe for detecting the progress of successive extension reaction cycles. Signal detection is dependent upon DNA synthesis and can occur with or without probe cleavage.
Owner:INTEGRATED DNA TECHNOLOGIES
High fidelity DNA polymerase compositions and uses therefor
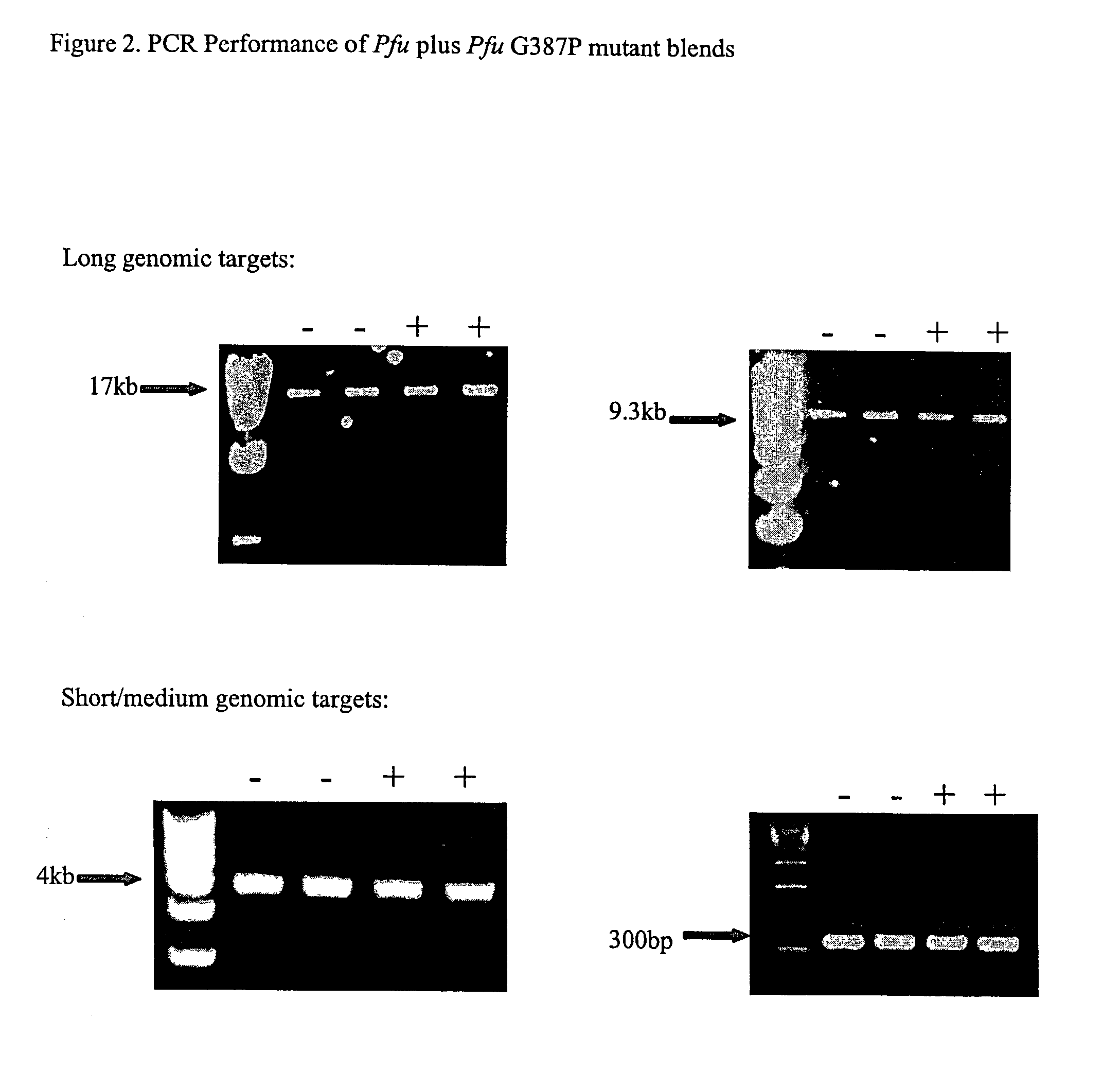
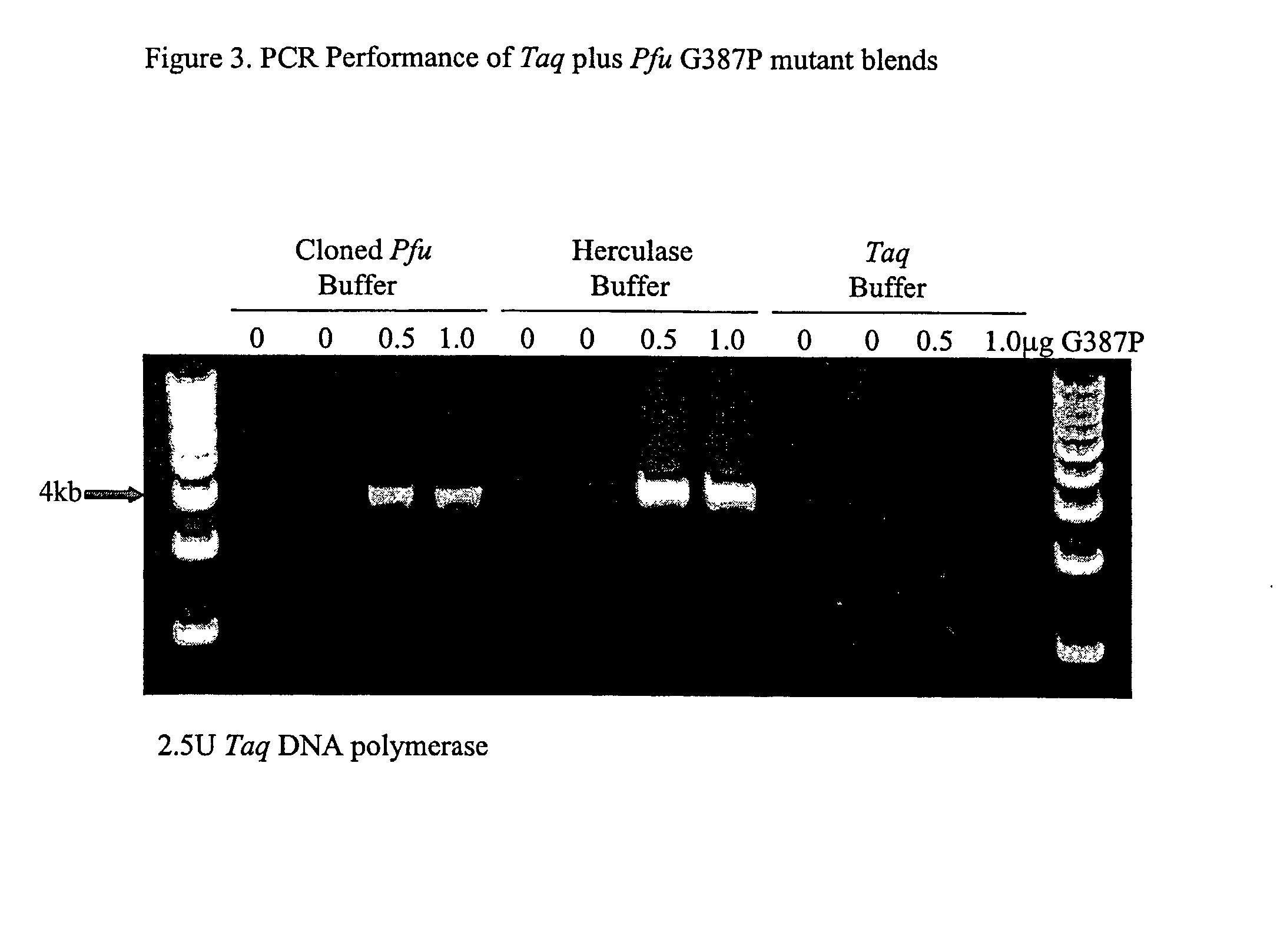
The subject invention relates to compositions comprising an enzyme mixture which comprises a first enzyme and a second enzyme, where the first enzyme comprises a DNA polymerization activity and the second enzyme comprises an 3′-5′ exonuclease activity and a reduced DNA polymerization activity. The invention also relates to the above compositions in kit format and methods for high fidelity DNA synthesis using the subject compositions of the invention.
Owner:AGILENT TECH INC
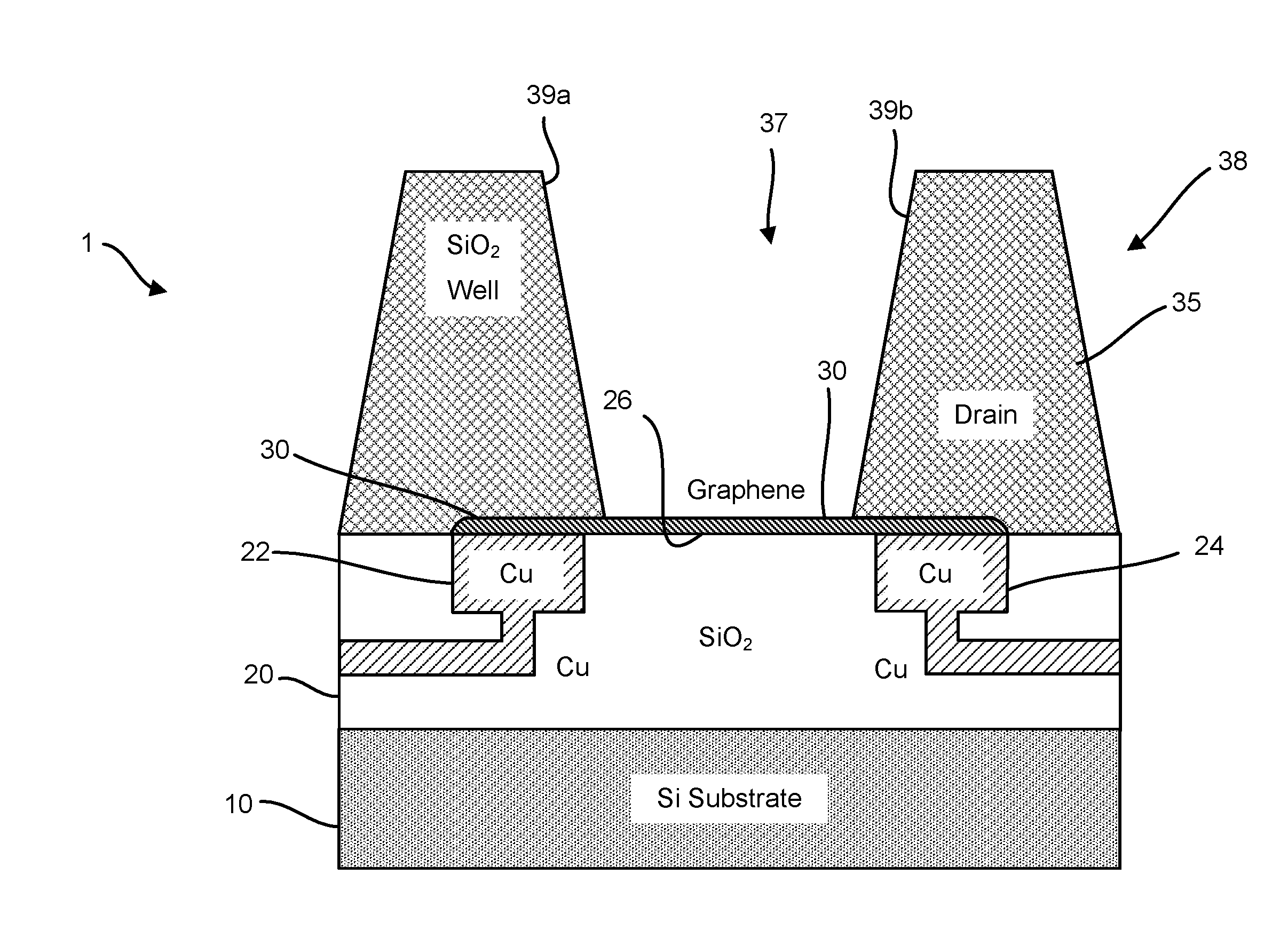
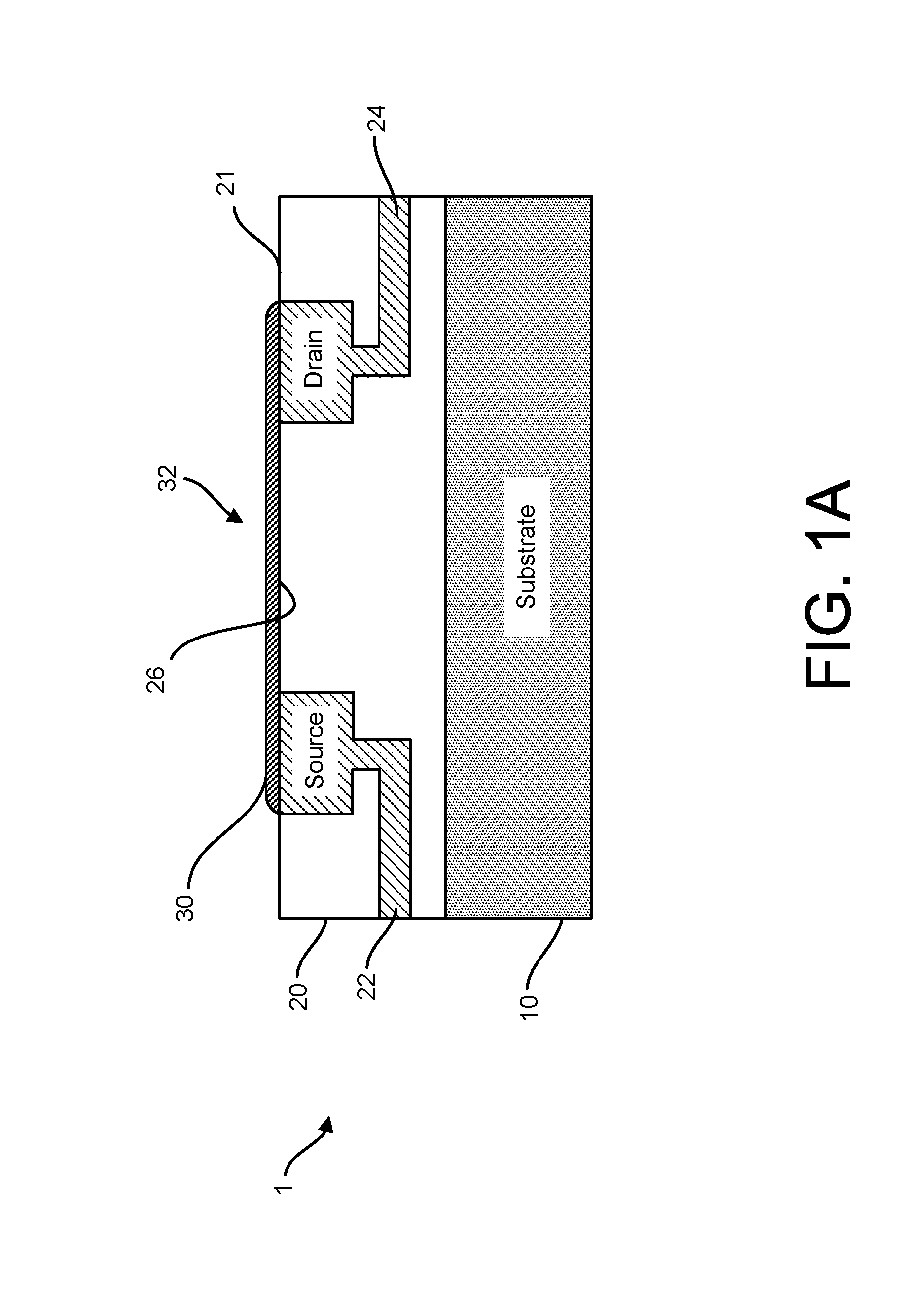
Graphene fet devices, systems, and methods of using the same for sequencing nucleic acids
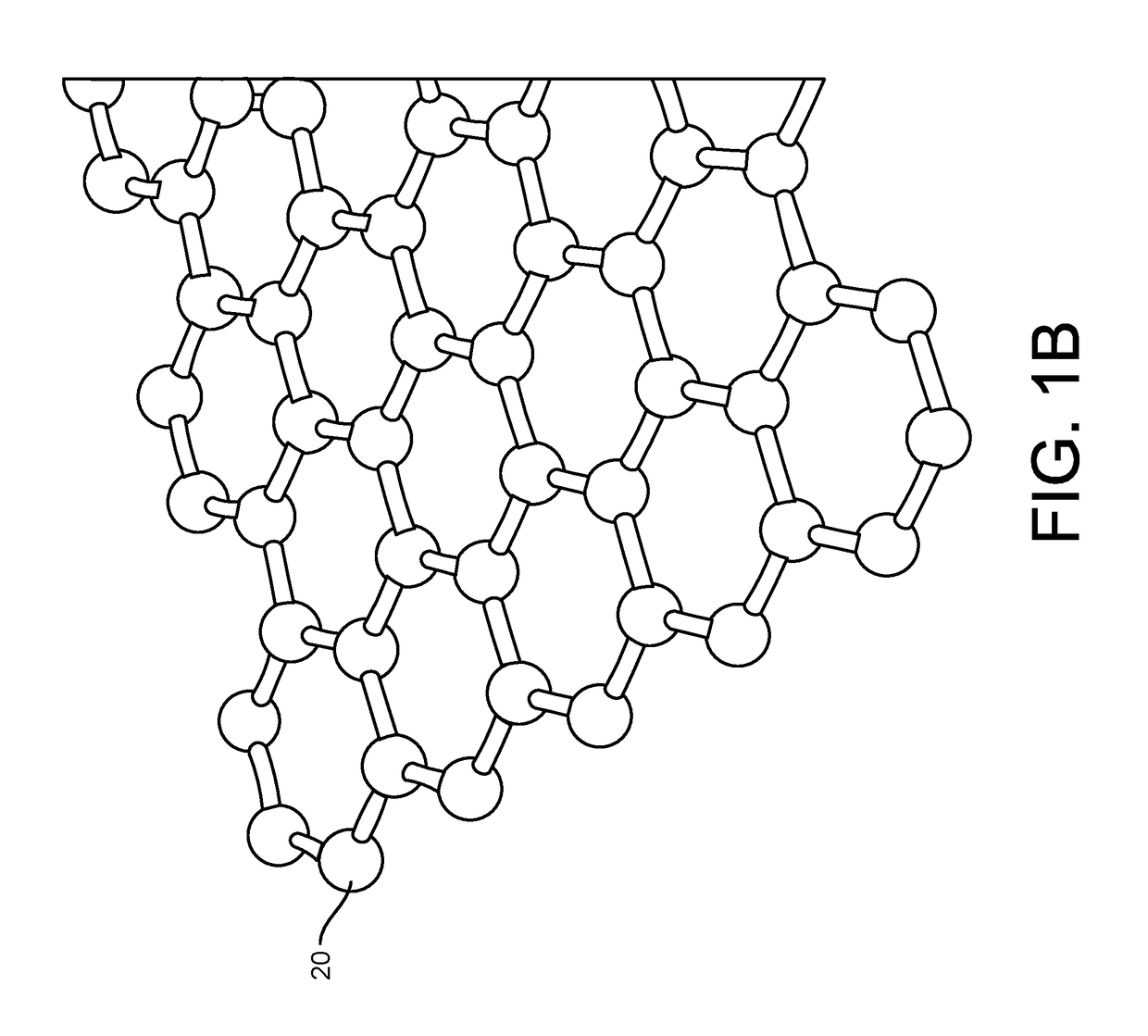
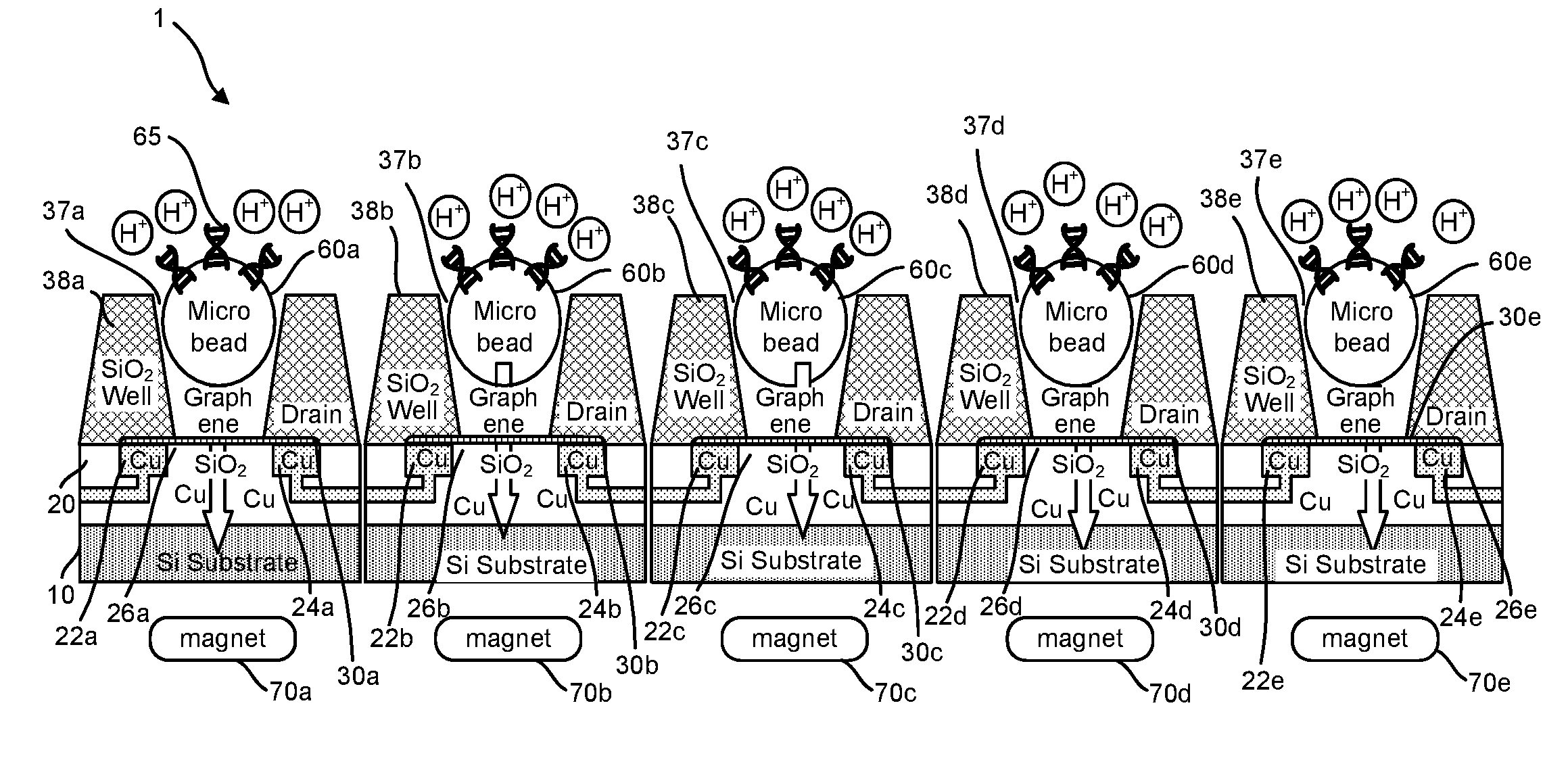
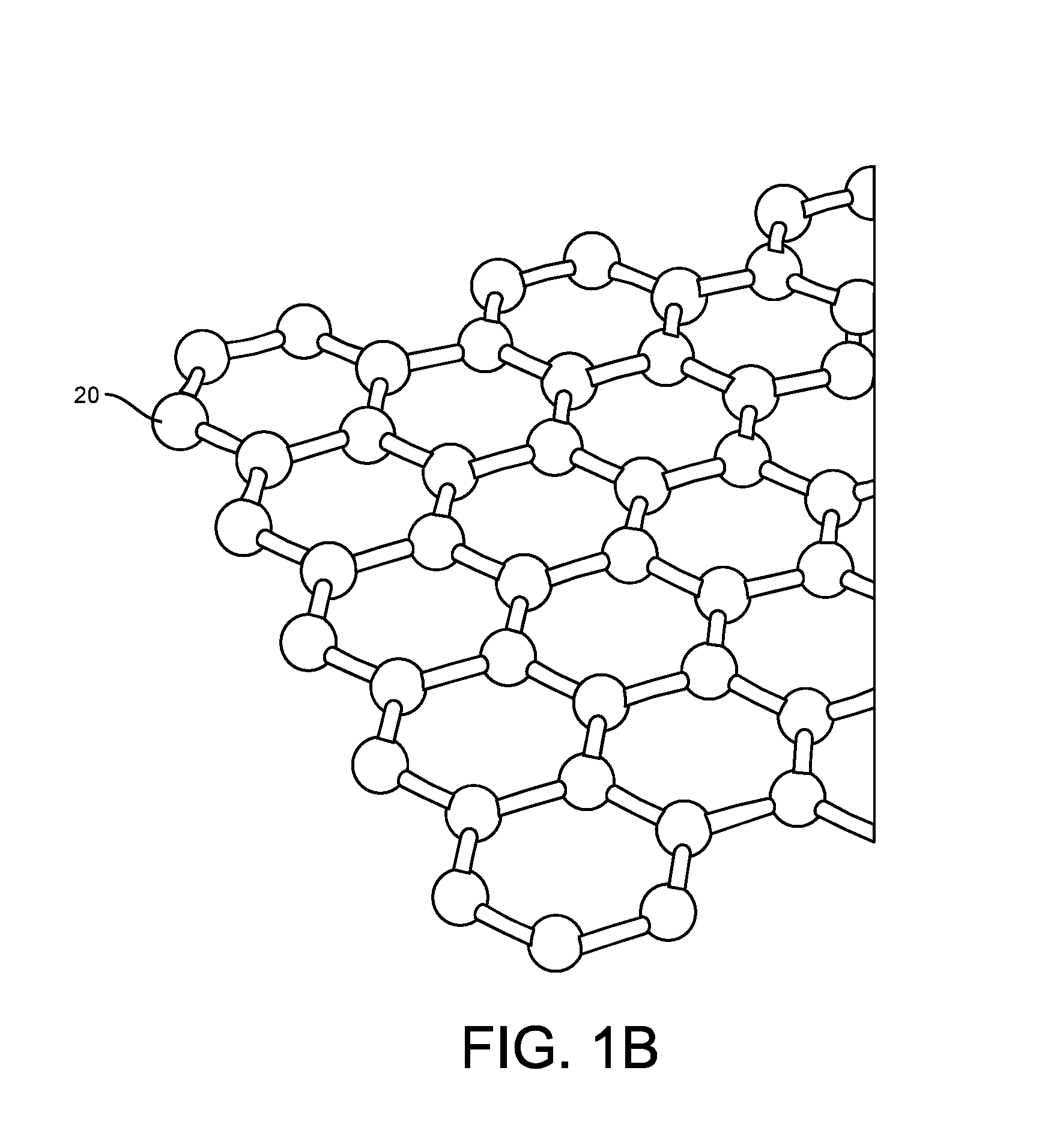
ActiveUS20160265047A1Number of EliminationsFaster data acquisitionTransistorMicrobiological testing/measurementSensor arrayReaction layer
Provided herein are devices, systems, and methods of employing the same for the performance of bioinformatics analysis. The apparatuses and methods of the disclosure are directed in part to large scale graphene FET sensors, arrays, and integrated circuits employing the same for analyte measurements. The present GFET sensors, arrays, and integrated circuits may be fabricated using conventional CMOS processing techniques based on improved GFET pixel and array designs that increase measurement sensitivity and accuracy, and at the same time facilitate significantly small pixel sizes and dense GFET sensor based arrays. Improved fabrication techniques employing graphene as a reaction layer provide for rapid data acquisition from small sensors to large and dense arrays of sensors. Such arrays may be employed to detect a presence and / or concentration changes of various analyte types in a wide variety of chemical and / or biological processes, including DNA hybridization and / or sequencing reactions. Accordingly, GFET arrays facilitate DNA sequencing techniques based on monitoring changes in hydrogen ion concentration (pH), changes in other analyte concentration, and / or binding events associated with chemical processes relating to DNA synthesis within a gated reaction chamber of the GFET based sensor.
Owner:CARDEA BIO INC
Encoding method and decoding method for performing information storage by means of DNA
InactiveCN105022935ACoding method is simple and easyConsider efficiencySpecial data processing applicationsDecoding methodsMagnetic media
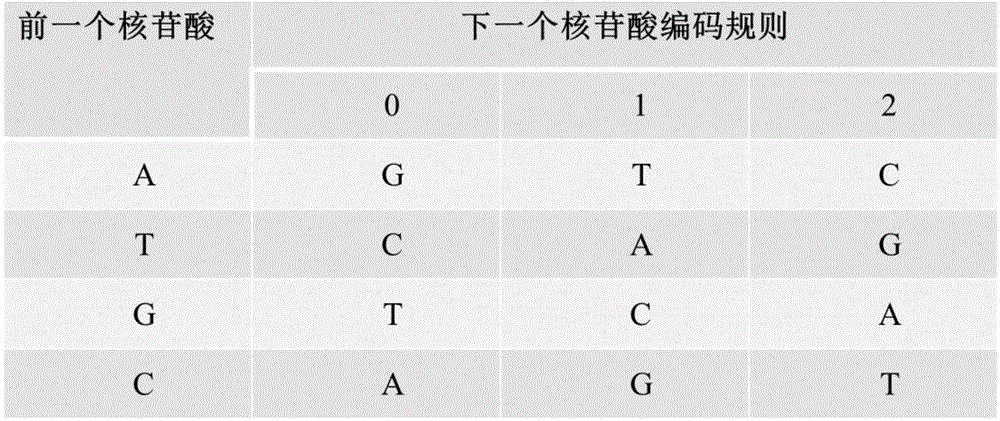
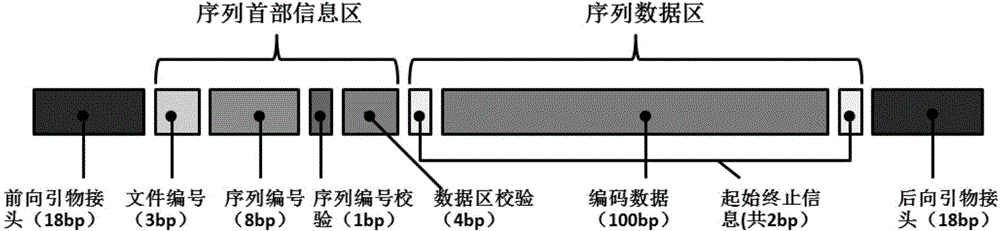
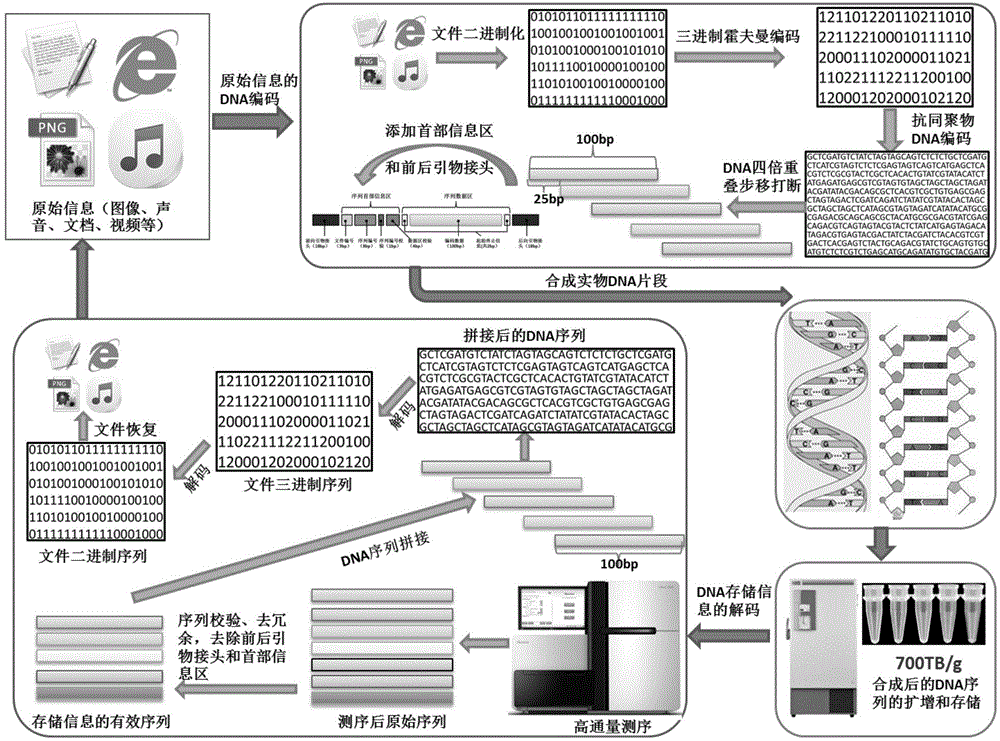
The present invention relates to an encoding method and an decoding method for performing information storage by means of DNA. Different from a conventional computer magnetic medium, an information write-in mode of DNA storage is that after information is encoded, an oligonucleotide chain with a certain length is synthesized by utilizing an oligonucleotide chain synthesis technology and the synthesized oligonucleotide chain is stored in a powder form; and a reading technology of DNA storage is that the oligonucleotide chain is sequenced by utilizing a high-throughput sequencing technology and after being spliced, sequenced fragments are transcoded, so that an initial computer multimedia file can be restored. Due to the characteristics of the DNA oligonucleotide chain, in the design of an encoding mode, a random error possibly existing in the DNA synthesis and sequencing process can be taken into full consideration, and error authentication and multiple cover segment are performed on the DNA fragments. The encoding method for a DNA storage technology, which is constructed by the present invention, is simple, convenient and easy to operate and can be applied to transform the computer multimedia files in various formats into DNA sequences so as to perform information storage.
Owner:QINGDAO INST OF BIOENERGY & BIOPROCESS TECH CHINESE ACADEMY OF SCI
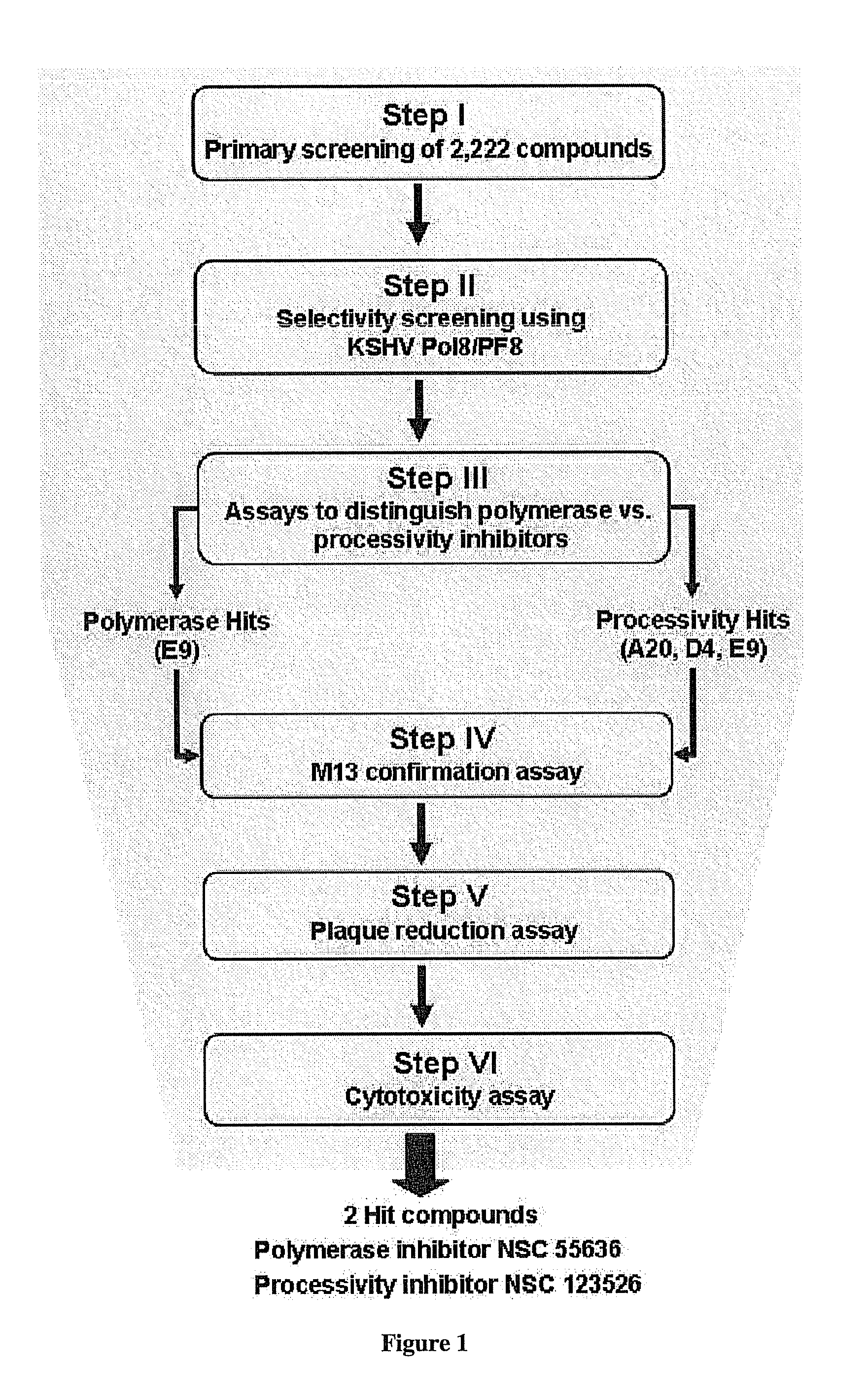
Therapeutic compounds for blocking DNA synthesis of pox viruses
ActiveUS20100035887A1Inhibition of replicationInhibit and reduce activityBiocideTetracycline active ingredientsMedicinePoxvirus Infections
This invention provides methods of inhibiting replication of a poxvirus by contacting a poxvirus with a compound having formula I, formula XXI, formula XXXII, or formula XLI which in turn reduce, inhibit, or abrogate poxvirus DNA polymerase activity and / or its interaction with its processivity factor. Formula I, formula XXI, formula XXXII, or formula XLI can be utilized to treat humans and animals suffering from a poxvirus infection. Pharmaceutical compositions for treating poxvirus infected subjects are also provided.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
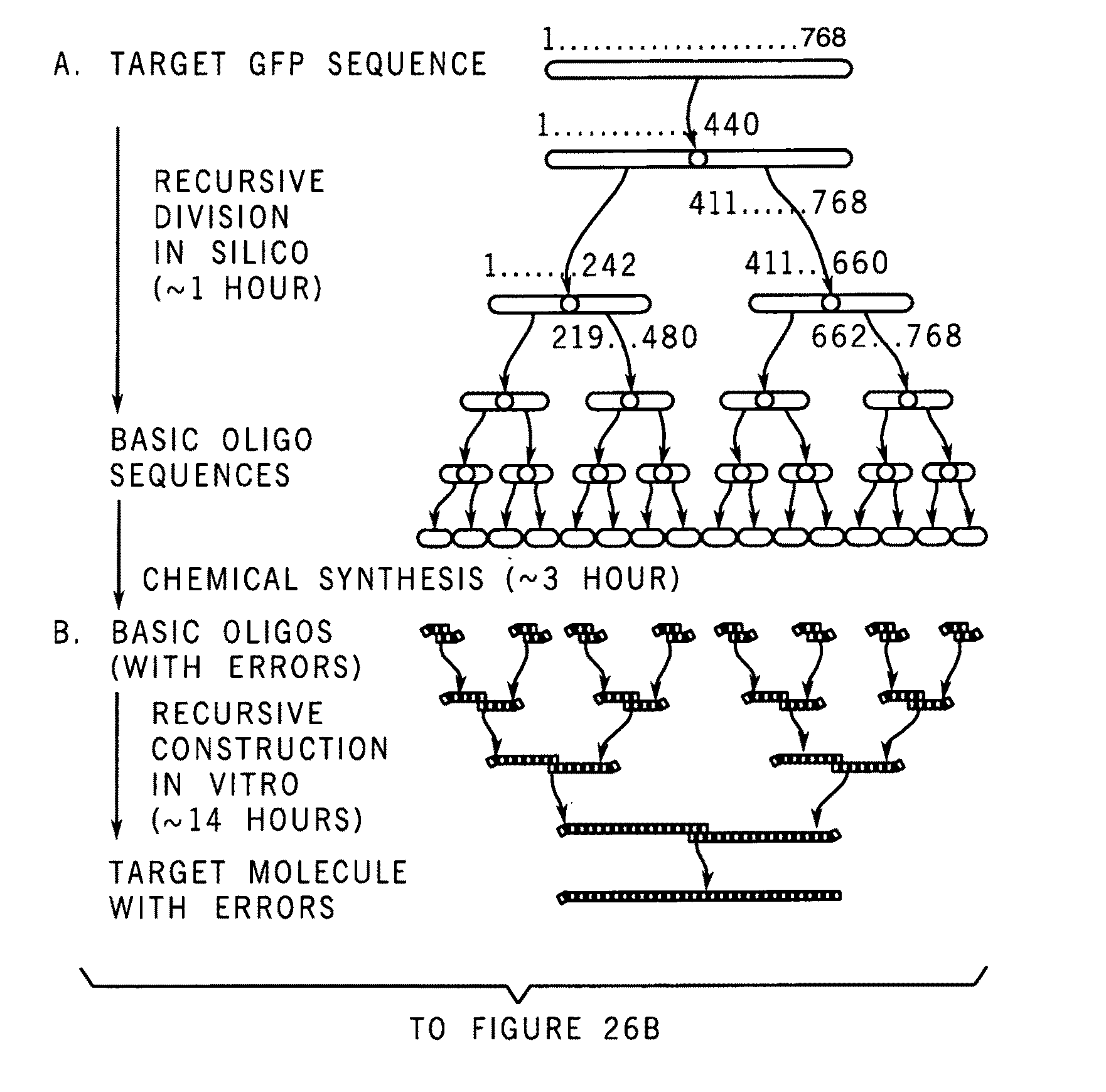
Programmable iterated elongation: a method for manufacturing synthetic genes and combinatorial DNA and protein libraries
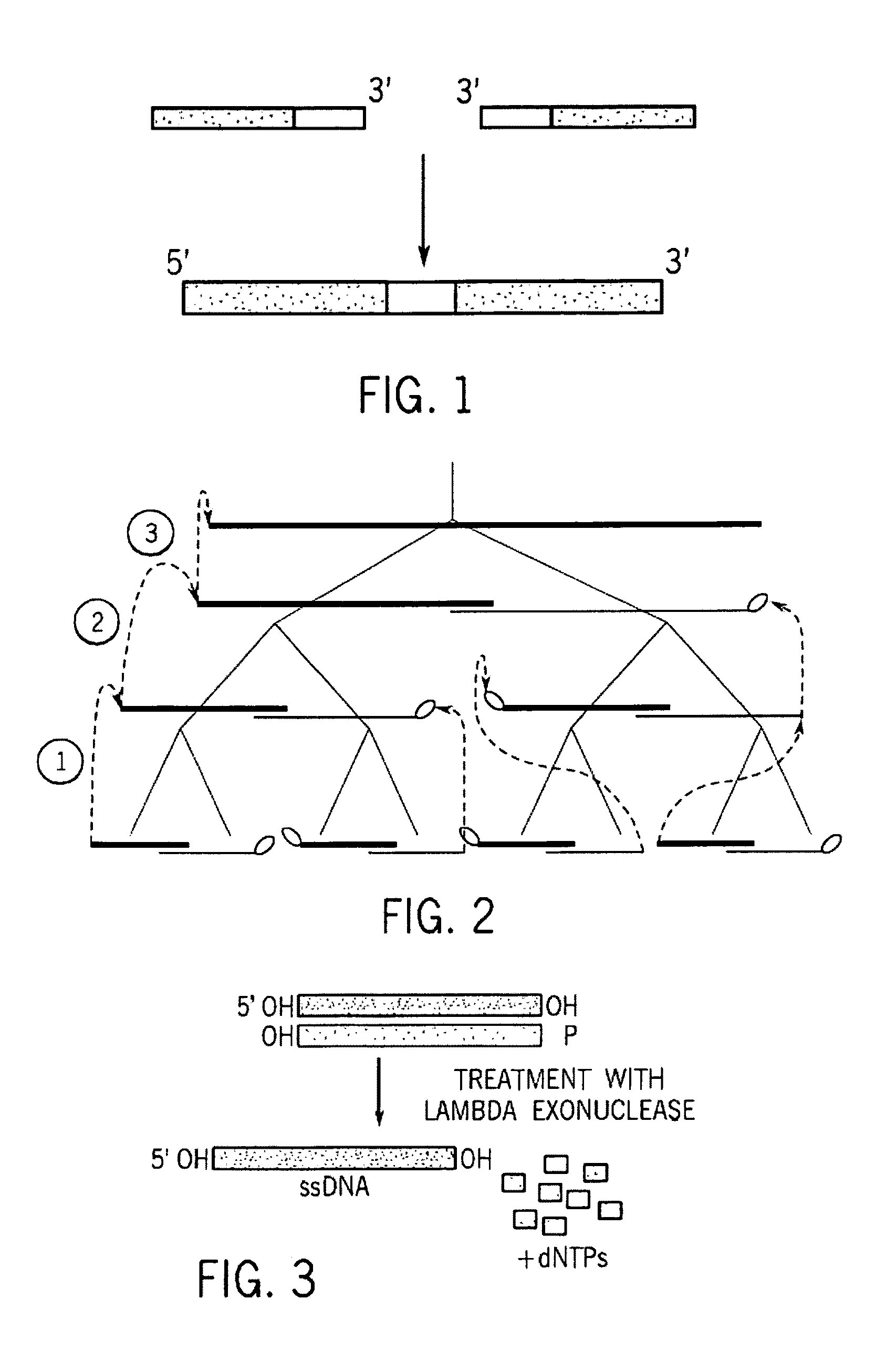
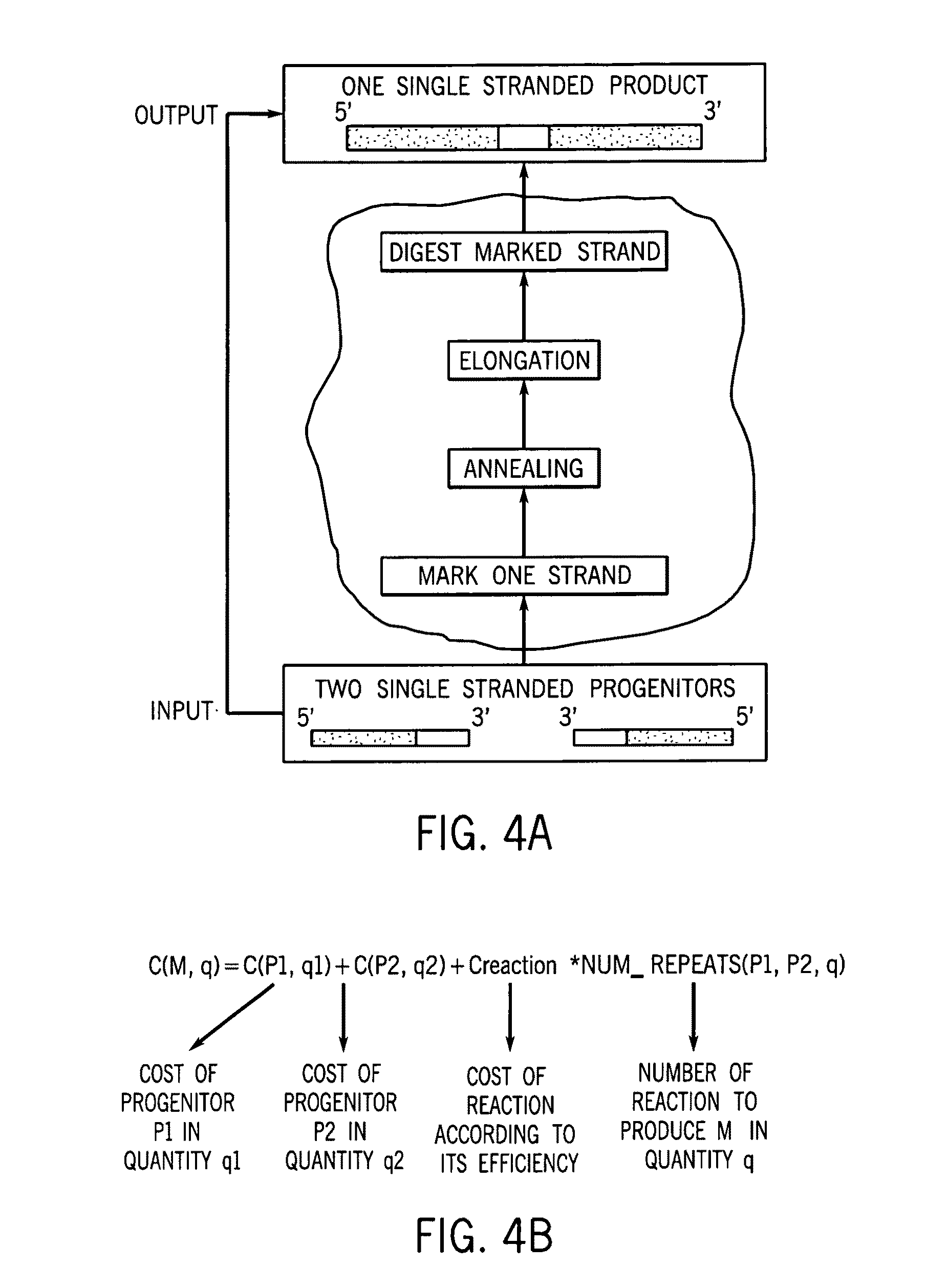
A method for manufacturing synthetic genes and combinatorial DNA and protein libraries, termed here Divide and Conquer-DNA synthesis (D&C-DNA synthesis) method. The method can be used in a systematic and automated way to synthesize any long DNA molecule and, more generally, any combinatorial molecular library having the mathematical property of being a regular set of strings. The D&C-DNA synthesis method is an algorithm design paradigm that works by recursively breaking down a problem into two or more sub-problems of the same type. The division of long DNA sequences is done in silico. The assembly of the sequence is done in vitro. The D&C-DNA synthesis method protocol consists of a tree, in which each node represents an intermediate sequence. The internal nodes are created in elongation reactions from their daughter nodes, and the leaves are synthesized directly. After each elongation only one DNA strand passes to the next level in the tree until receiving the final product. Optionally and preferably, error correction is performed to correct any errors which may have occurred during the synthetic process.
Owner:YEDA RES & DEV CO LTD
Graphene FET devices, systems, and methods of using the same for sequencing nucleic acids
ActiveUS9618474B2Faster data acquisitionImprove accuracyTransistorMicrobiological testing/measurementReaction layerSensor array
Provided herein are devices, systems, and methods of employing the same for the performance of bioinformatics analysis. The apparatuses and methods of the disclosure are directed in part to large scale graphene FET sensors, arrays, and integrated circuits employing the same for analyte measurements. The present GFET sensors, arrays, and integrated circuits may be fabricated using conventional CMOS processing techniques based on improved GFET pixel and array designs that increase measurement sensitivity and accuracy, and at the same time facilitate significantly small pixel sizes and dense GFET sensor based arrays. Improved fabrication techniques employing graphene as a reaction layer provide for rapid data acquisition from small sensors to large and dense arrays of sensors. Such arrays may be employed to detect a presence and / or concentration changes of various analyte types in a wide variety of chemical and / or biological processes, including DNA hybridization and / or sequencing reactions. Accordingly, GFET arrays facilitate DNA sequencing techniques based on monitoring changes in hydrogen ion concentration (pH), changes in other analyte concentration, and / or binding events associated with chemical processes relating to DNA synthesis within a gated reaction chamber of the GFET based sensor.
Owner:CARDEA BIO INC
Graphene fet devices, systems, and methods of using the same for sequencing nucleic acids
ActiveUS20170018626A1Number of EliminationsFaster data acquisitionTransistorMicrobiological testing/measurementReaction layerSensor array
Provided herein are devices, systems, and methods of employing the same for the performance of bioinformatics analysis. The apparatuses and methods of the disclosure are directed in part to large scale graphene FET sensors, arrays, and integrated circuits employing the same for analyte measurements. The present GFET sensors, arrays, and integrated circuits may be fabricated using conventional CMOS processing techniques based on improved GFET pixel and array designs that increase measurement sensitivity and accuracy, and at the same time facilitate significantly small pixel sizes and dense GFET sensor based arrays. Improved fabrication techniques employing graphene as a reaction layer provide for rapid data acquisition from small sensors to large and dense arrays of sensors. Such arrays may be employed to detect a presence and / or concentration changes of various analyte types in a wide variety of chemical and / or biological processes, including DNA hybridization and / or sequencing reactions. Accordingly, GFET arrays facilitate DNA sequencing techniques based on monitoring changes in hydrogen ion concentration (pH), changes in other analyte concentration, and / or binding events associated with chemical processes relating to DNA synthesis within a gated reaction chamber of the GFET based sensor.
Owner:CARDEA BIO INC
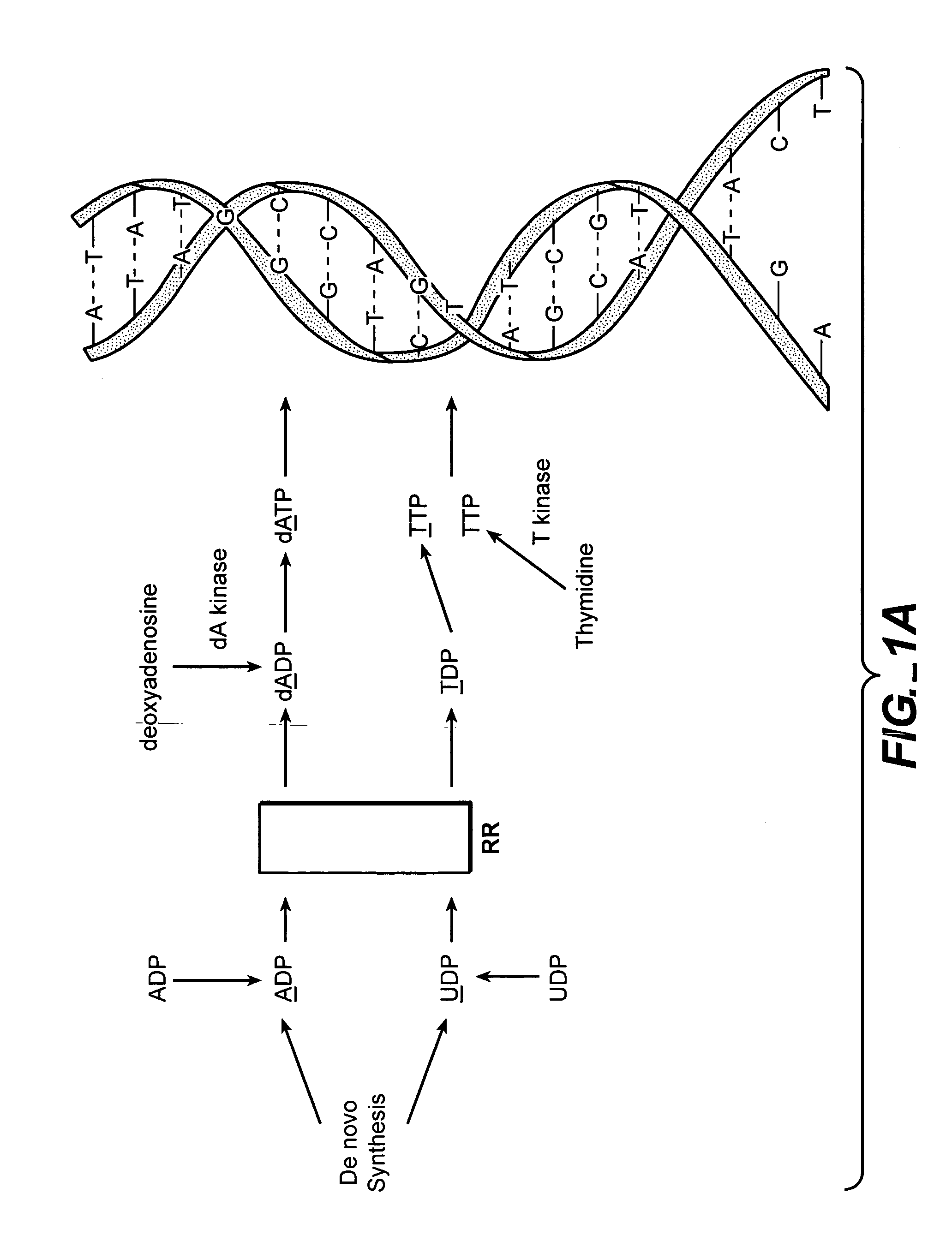
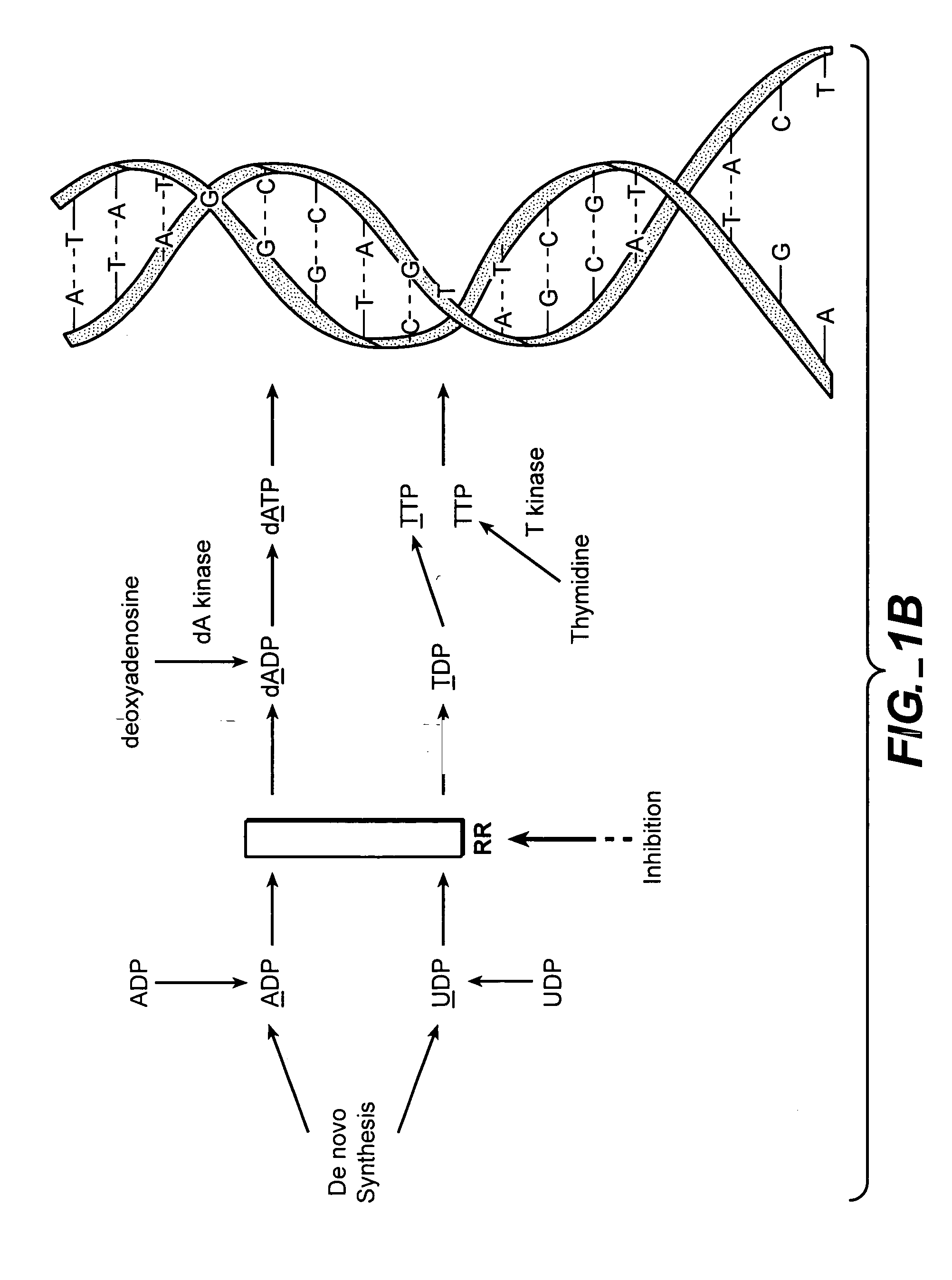
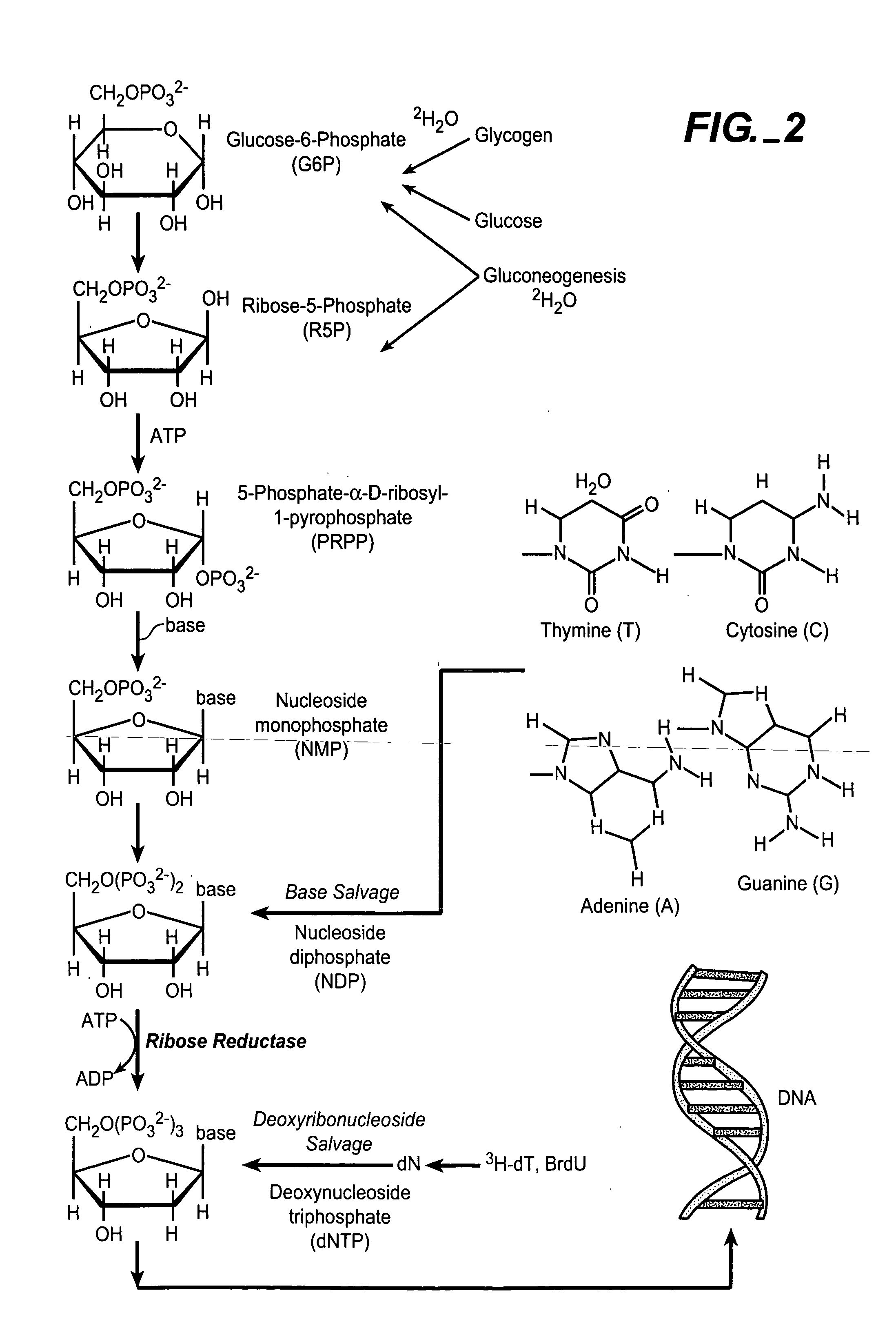
In vivo measurement of the relative fluxes through ribonucleotide reductase vs. deoxyribonucleoside pathways using isotopes
InactiveUS20050255509A1Increase salvageHigh activityCompound screeningApoptosis detectionRate-determining stepDeoxyribonucleotide biosynthesis
The methods of the present invention allow for the measurement of ribonucleotide reductase (RR) activity, an important enzyme in the de novo DNA synthesis pathway. Ribonucleotide reductase converts all four ribonucleotides to their deoxy form and is a rate-controlling step in this pathway. Biosynthetic pathways of deoxyribonucleotides (dN) have received considerable attention in the context of anti-proliferative chemotherapy. Inhibitors of various steps in dN biosynthesis, including inhibitors of RR are among the most useful chemotherapeutic agents in cancer, viral infections, and other therapeutic uses. DNA synthesis from the dN salvage pathway is also an important component to DNA replication. The relative contributions from RR vs. salvage pathways are critical to the actions and effectiveness of chemotherapeutic agents that act on nucleoside metabolic pathways. Until now, however, it has not been possible to study these metabolic processes in vivo. Disclosed within are methods of measuring RR activity in vivo and in vitro which find use, among other things, in drug discovery, development, and approval.
Owner:KINEMED
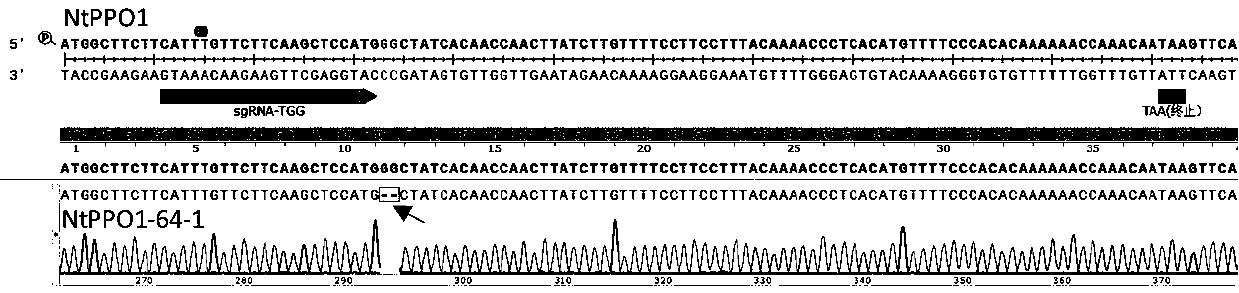
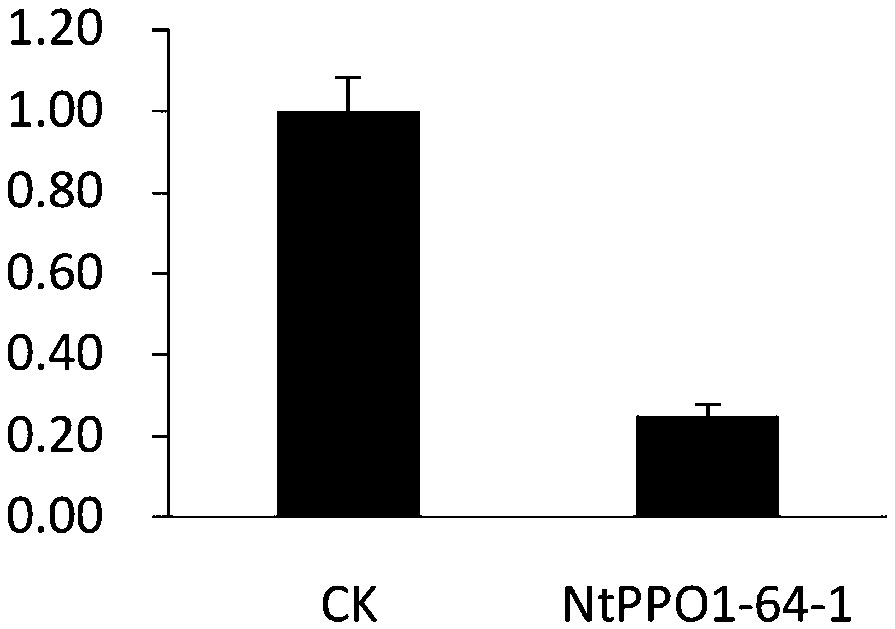
Tobacco polyphenol oxidase gene NtPPO1 and site-directed mutagenesis method and application thereof
InactiveCN107653256AReduce expressionReduce PPO enzyme activityHydrolasesOxidoreductasesNicotiana tabacumPolyphenol oxidase
The invention discloses a tobacco polyphenol oxidase gene NtPPO1 and a site-directed mutagenesis method and application thereof. The nucleotide sequence of the tobacco polyphenol oxidase gene NtPPO1 is shown in SEQ ID:No.1, the coded amino acid sequence is shown in SEQ ID:No.2. The invention further discloses a clone method of the tobacco polyphenol oxidase gene NtPPO1. The clone method comprisesthe specific steps that firstly, cDNAs of tobacco leaves are synthesized; secondly, PCR amplification of the NtPPO1 gene is conducted; thirdly, a CRISPR / Cas9 carrier of the NtPPO1 gene is constructed;fourthly, sequencing detection of the NtPPO1 gene mutation is conducted. The NtPPO1 gene has a wide application prospect in preventing tobacco browning. The tobacco polyphenol oxidase gene and the encoded protein thereof provide the gene and technology support for crops especially for anti-browning breeding of tobaccos.
Owner:YUNNAN ACAD OF TOBACCO AGRI SCI
Specific target polypeptide self-assembled nano-carrier, drug-carrying nanoparticle and preparation method
ActiveCN106822036AGrowth inhibitionInhibit synthesisOrganic active ingredientsMacromolecular non-active ingredientsNanocarriersBiocompatibility Testing
The invention relates to the technical field of biology and in particular relates to a specific target polypeptide self-assembled nanoparticle. The nanoparticle is prepared from a hydrophobic anti-tumor drug with a therapeutic dosage and an amphipathic polypeptide which covers the periphery of the hydrophobic anti-tumor drug through a self-assembling manner, wherein the amphipathic polypeptide is a target peptide which can be used for specifically targeting a epidermal growth factor receptor of a tumor cell; a terminal N of the target peptide is coupled with a hydrophobic functional molecule. After the nanoparticle targets the tumor cell, the target peptide is exposed; the nanoparticle targets the tumor cell and enters the tumor cell through receptor-mediated endocytosis, and the drug is released to inhibit DNA (Deoxyribonucleic Acid) synthesis and repairing; the nanoparticle has dual killing effects on the tumor cells and the growth of the tumor cell is inhibited. The amphipathic polypeptide does not generate a covalent bond in a self-assembling process and no reverse reaction is caused; the specific target polypeptide self-assembled nanoparticle is used for treating tumors and has the advantages of no toxin and good biocompatibility.
Owner:THE NAT CENT FOR NANOSCI & TECH NCNST OF CHINA
Kits for amplifying DNA
ActiveUS8183359B2Reduce appearance problemsMinimize and substantially eliminate emergenceSugar derivativesMicrobiological testing/measurementAmplification dnaA-DNA
Kits for amplifying DNA which include a priming oligonucleotide that hybridizes to a 3′-end of a DNA target sequence, a displacer oligonucleotide that hybridizes to a target nucleic acid containing the DNA target sequence at a position upstream from the priming oligonucleotide, and a promoter oligonucleotide that includes a region that hybridizes to a 3′-region of a DNA primer extension product that includes the priming oligonucleotide and a promoter for an RNA polymerase. The priming oligonucleotide does not include an RNA region that hybridizes to the target nucleic acid and is selectively degraded by an enzyme activity when hybridized to the target nucleic acid. The kits do not include a restriction endonuclease and oligonucleotides that include a promoter for an RNA polymerase are all modified to prevent the initiation of DNA synthesis therefrom.
Owner:GEN PROBE INC
Reversible terminal and synthesis and use in DNA synthesis sequencing thereof
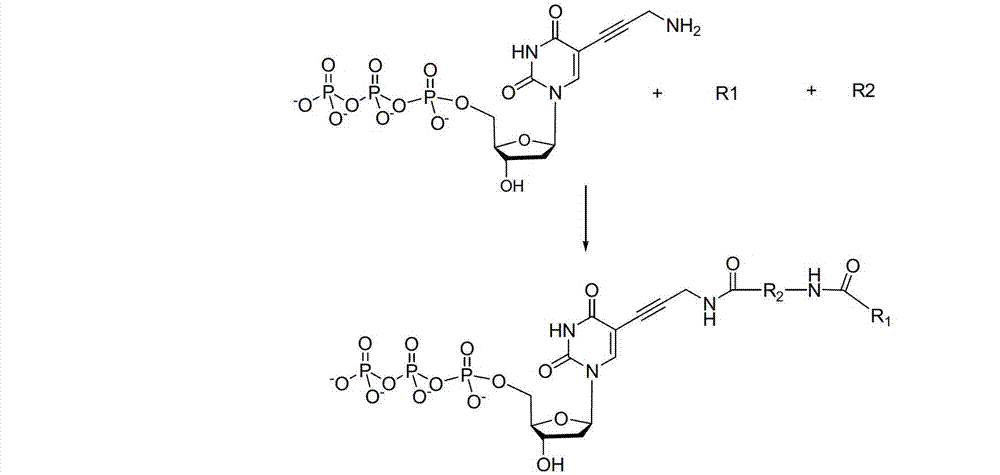
ActiveCN103087131ARaw materials are easy to obtainSugar derivativesMicrobiological testing/measurementChemical reactionHigh energy
The invention discloses a reversible terminal and synthesis and use in DNA synthesis sequencing thereof. The structural formula of the reversible terminal is described according to formula (I), wherein R1 is fluorescein and R2 is a connection unit. The cracking reversible terminal of the invention is available for DNA synthesis sequencing. At the same time, as synthesis required material is easily available and synthesis processes are all conventional chemical reactions, the reversible terminal disclosed by the invention is applicable for large-scale promotion and has good practical prospect due to a biological evaluation result showing that the reversible terminal is capable of totally satisfying biochemical reaction requirements of high-energy sequencing.
Owner:SHANGHAI JIAO TONG UNIV
Methods for amplifying polymeric nucleic acids
InactiveUS20050255486A1Avoid small quantitiesEfficiently generates extension productsSugar derivativesMicrobiological testing/measurementDouble strandedPolymer
The invention provides compositions and methods for amplifying nucleic acid polymer sequences in a high complexity nucleic acid sample. The unique compositions of the invention include a primer set composed of a mixture of two types of primers for DNA synthesis. For extension in one direction, the primers all contain modifications that destroy their ability to serve as templates that can be copied by DNA polymerases. For extension in the opposite direction the set includes at least one primer that can serve as a template and be replicated by DNA polymerases throughout its length. The method can be carried out by mixing the nucleic acid polymer sequence of interest with the set of DNA synthesis primers in an amplification reaction mixture. The reaction mixture is then subjected to temperature cycling analogous to the temperature cycling in PCR reactions. At least one primer in the primer set hybridizes to the nucleic acid polymer. It is preferred that the non-replicable primer hybridizes to the nucleic acid polymer and is extended to produce an extension product that contains sequence from the nucleic acid polymer to which the replicable primer then hybridizes. Of course, if the nucleic acid polymer is double stranded, both the replicable and nonreplicable primers will hybridize and be extended by DNA polymerase.
Owner:INTEGRATED DNA TECHNOLOGIES
Use of template switching for DNA synthesis
A method of preparing a DNA copy of a target polynucleotide using template switching is described. The method includes mixing a double stranded template / primer substrate made up of a DNA primer oligonucleotide associated with a complementary oligonucleotide template strand with a target polynucleotide in a reaction medium and adding a suitable amount of a non-retroviral reverse transcriptase to the reaction medium to extend the DNA primer oligonucleotide from its 3′ end to provide a DNA copy polynucleotide. The DNA copy polynucleotide includes a complementary target DNA polynucleotide that is synthesized using the target polynucleotide as a template. Methods of adding nucleotides to the double stranded template / primer substrate are also described. The method can be used to facilitate detection, PCR amplification, cloning, and determination of RNA and DNA sequences.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
Multiplexed genetic reporter assays and compositions
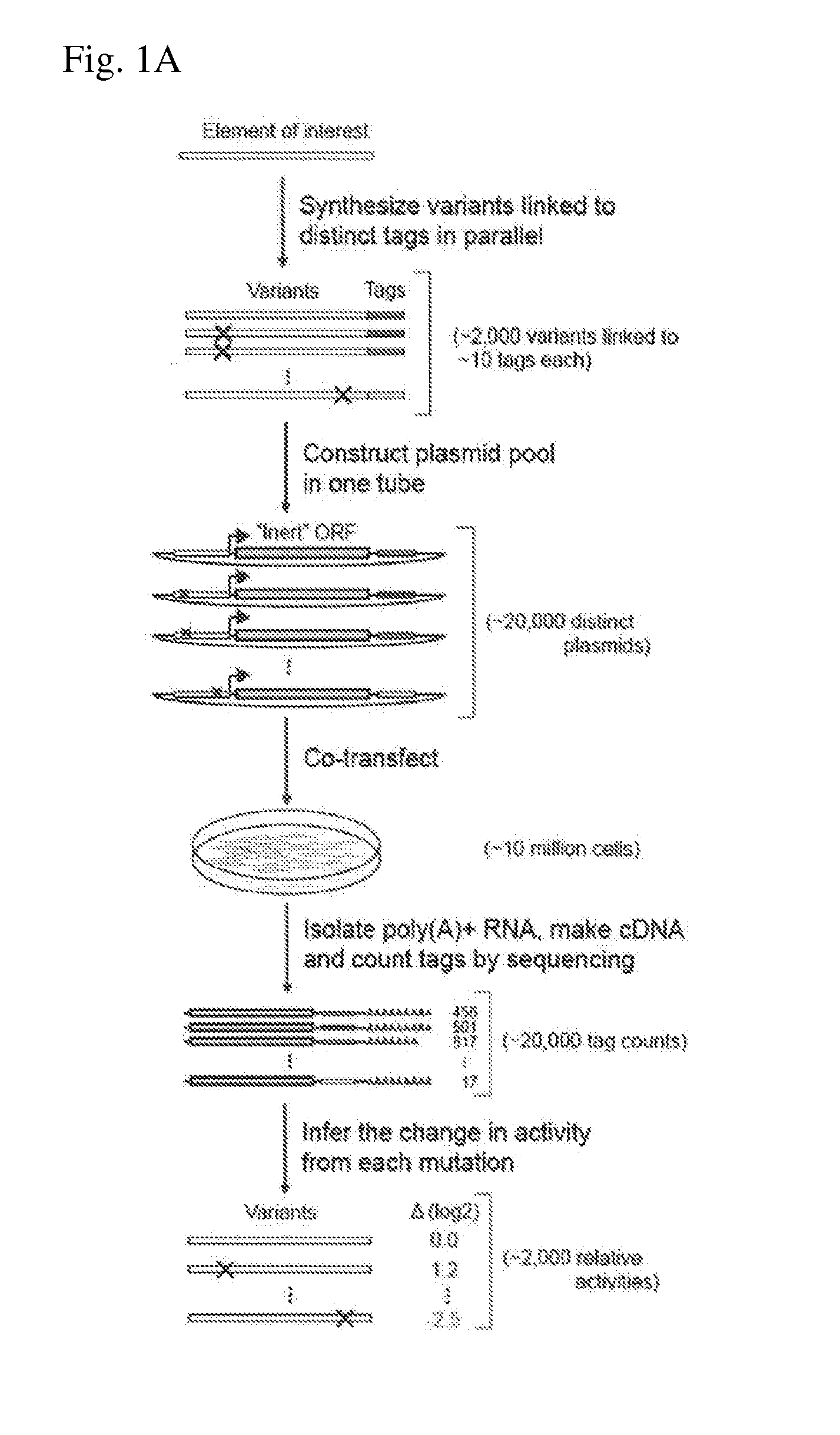
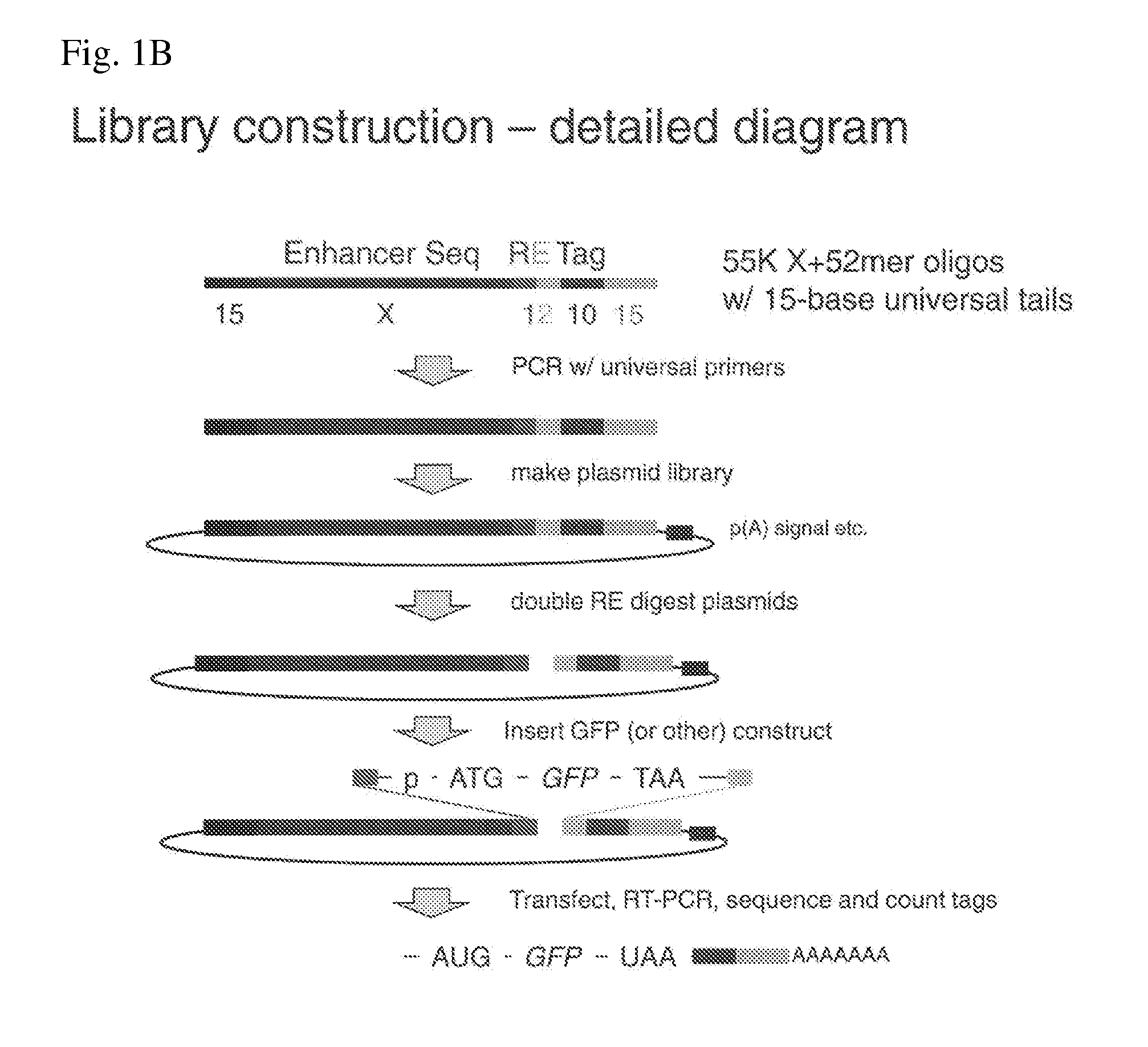
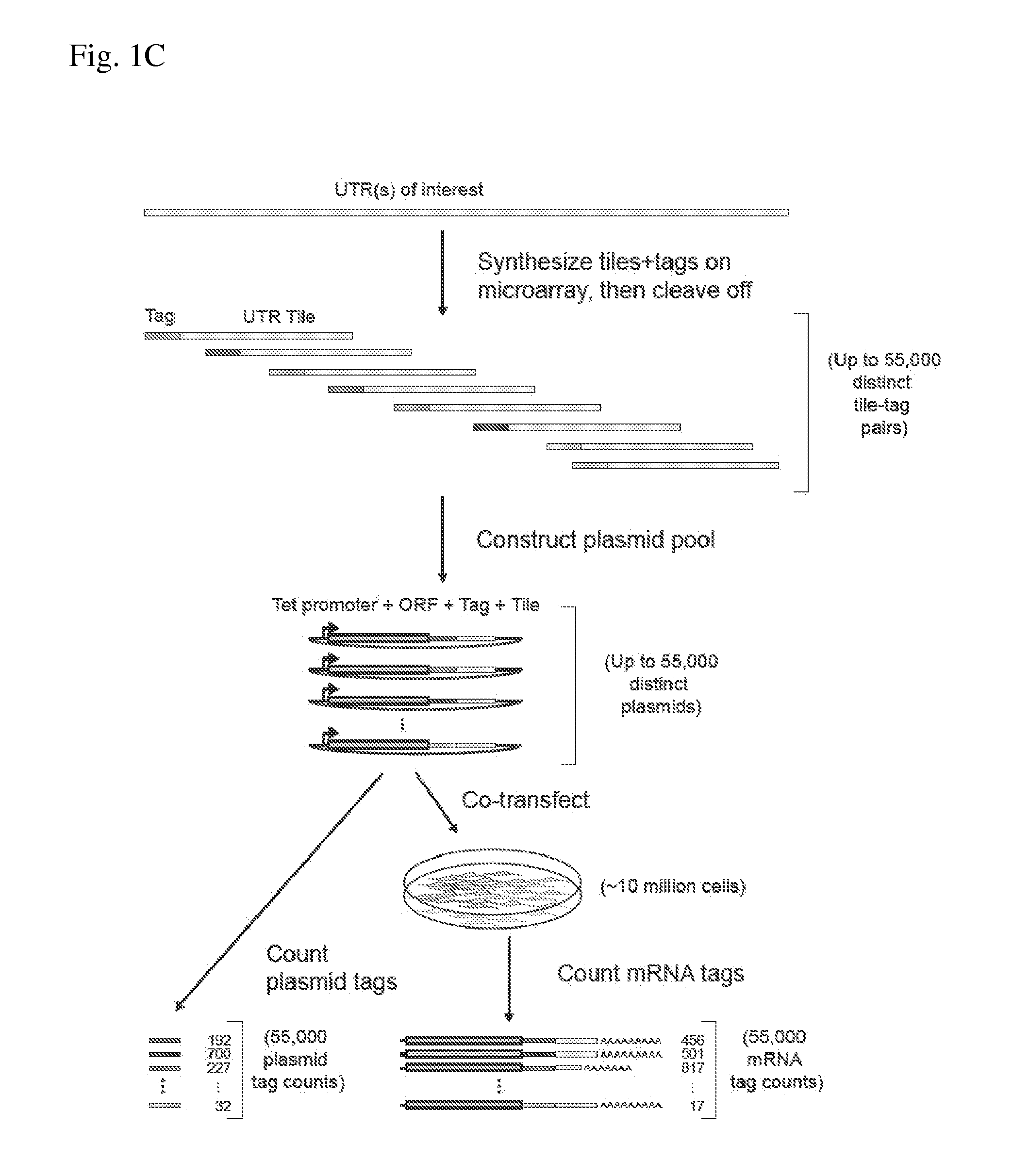
ActiveUS20140200163A1Easy to optimizeHigh resolutionMicrobiological testing/measurementLibrary screeningAssayTranscriptional regulation
The invention provides methods for determining the activity of a plurality of nucleic acid regulatory elements. These methods may facilitate, e.g., the systematic reverse engineering, and optimization of mammalian cis-regulatory elements at high resolution and at a large scale. The method may include integration of multiplexed DNA synthesis and sequencing technologies to generate and quantify the transcriptional regulatory activity of e.g., thousands of arbitrary DNA sequences in parallel in cell-based as says (e.g., mammalian cell based assays).
Owner:THE BROAD INST INC
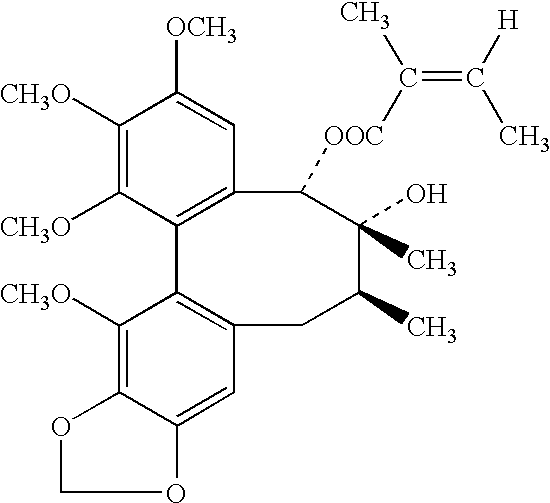
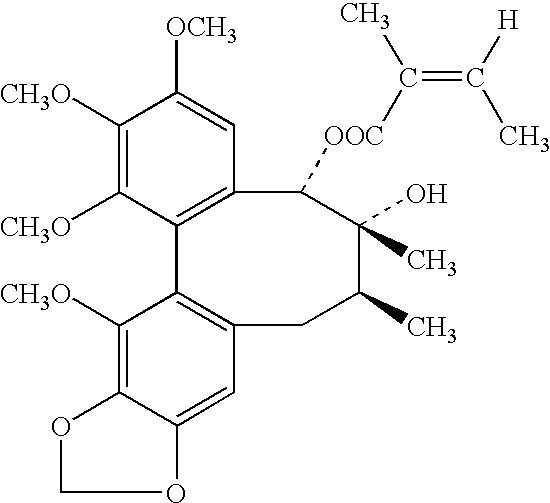
Plant drug for treatment of liver disease
The present invention related to safe plant drug for treatment of liver disease, specifically, this invention proves a safe plant drug Schisandrin and its preparation. Schisandrin has the following pharmaceutical functions: increasing tumor suppresson genes express activity, decreasing activity of oncogenes, increasing immune function, increasing liver DNA synthesis, decreasing serum alamine aminotransferase activity, increasing glutathione level, increasing glutathione reductase activity, decreasing lipid peroxidation of liver, increasing hepatic microsomal monooxygenases activity, increasing ATP content in liver, increasing energy metabolism activity, decreasing density lipoprotein oxidation, protecting gastrointestinal function, increasing killer cell activity, increasing complement activity, decreasing induced liver cancer activity and decreasing grown of cancer cells.
Owner:ZHAO XINXIAN
Methods for amplifying polymeric nucleic acids
InactiveUS20080038724A9Efficiently generates extension productsAvoid small quantitiesSugar derivativesMicrobiological testing/measurementDouble strandedPolymer
The invention provides compositions and methods for amplifying nucleic acid polymer sequences in a high complexity nucleic acid sample. The unique compositions of the invention include a primer set composed of a mixture of two types of primers for DNA synthesis. For extension in one direction, the primers all contain modifications that destroy their ability to serve as templates that can be copied by DNA polymerases. For extension in the opposite direction the set includes at least one primer that can serve as a template and be replicated by DNA polymerases throughout its length. The method can be carried out by mixing the nucleic acid polymer sequence of interest with the set of DNA synthesis primers in an amplification reaction mixture. The reaction mixture is then subjected to temperature cycling analogous to the temperature cycling in PCR reactions. At least one primer in the primer set hybridizes to the nucleic acid polymer. It is preferred that the non-replicable primer hybridizes to the nucleic acid polymer and is extended to produce an extension product that contains sequence from the nucleic acid polymer to which the replicable primer then hybridizes. Of course, if the nucleic acid polymer is double stranded, both the replicable and nonreplicable primers will hybridize and be extended by DNA polymerase.
Owner:INTEGRATED DNA TECHNOLOGIES
Modified triple-helix forming oligonucleotides for targeted mutagenesis
ActiveUS8658608B2Improve stabilityPeptide/protein ingredientsMammal material medical ingredientsBase JNucleotide
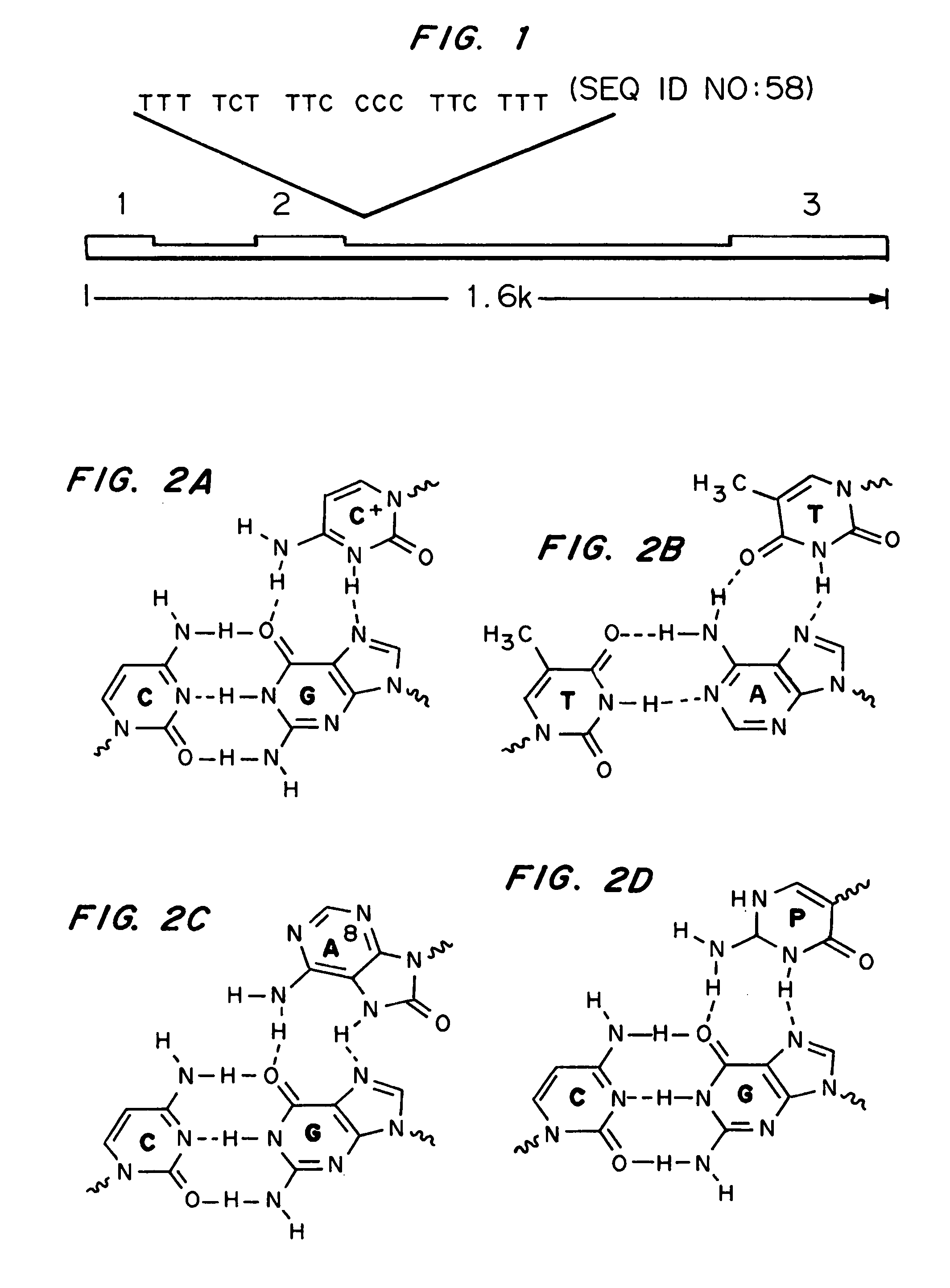
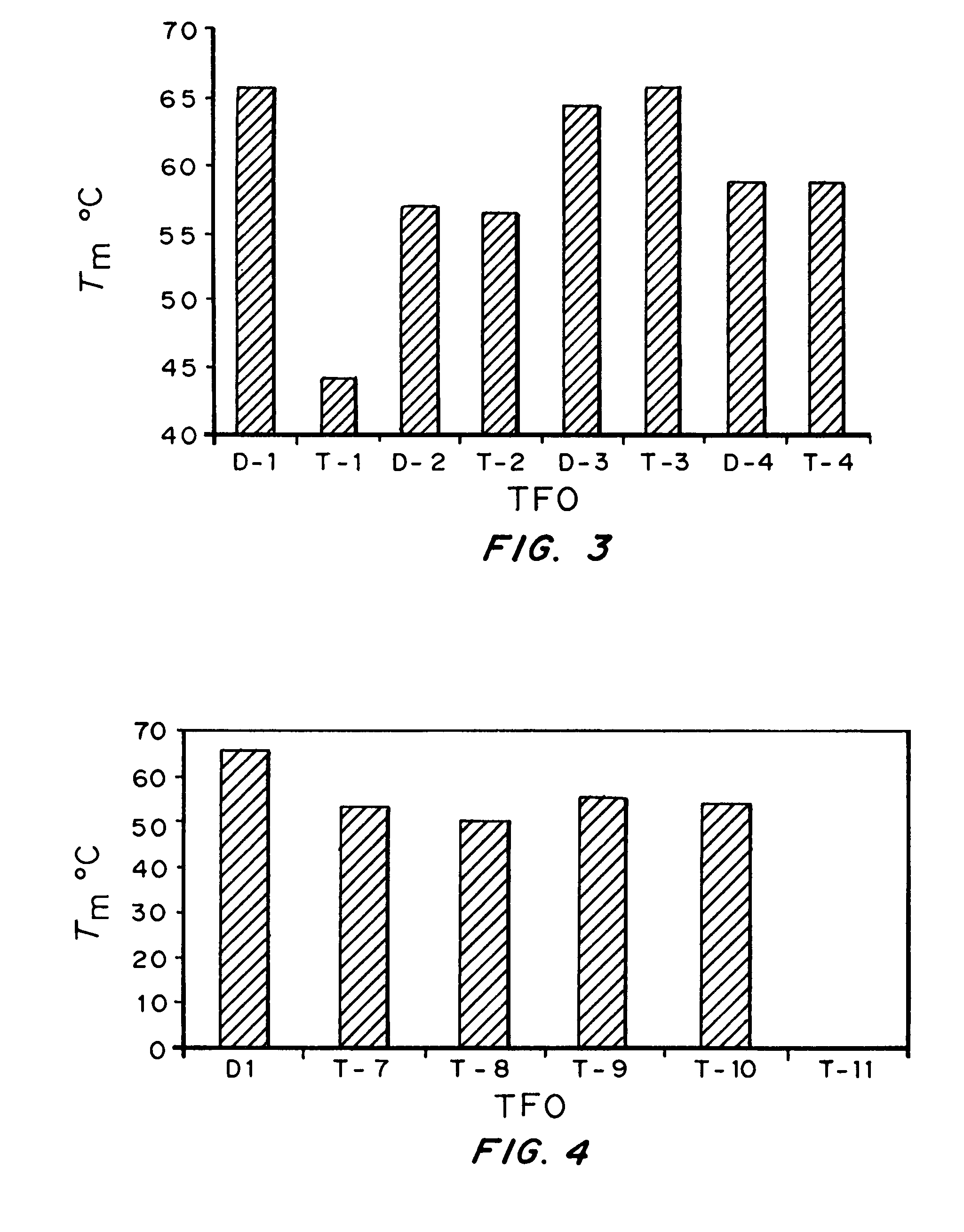
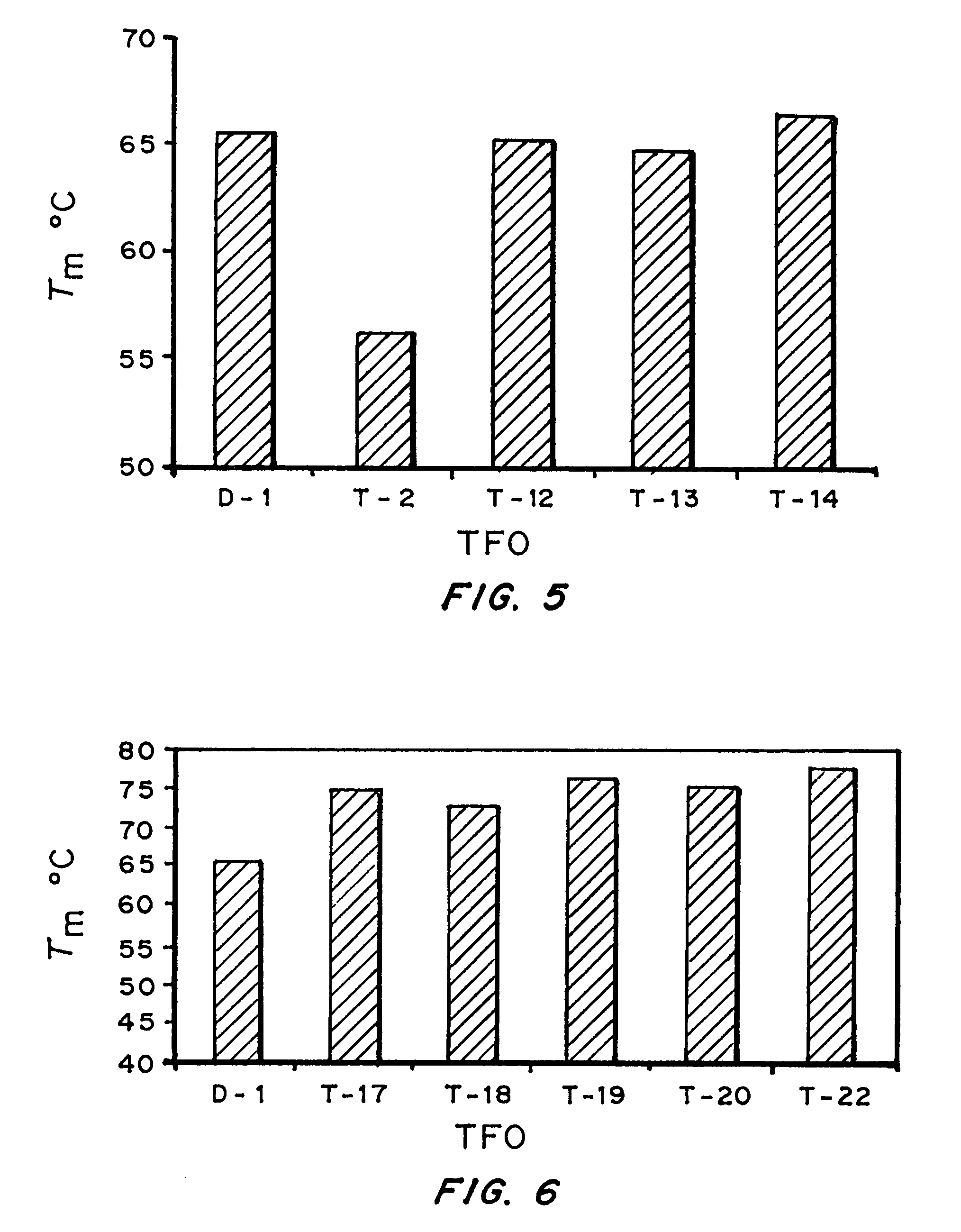
High affinity, chemically modified triplex-forming oligonucleotides (TFOs) and methods for use thereof are disclosed. TFOs are defined as triplex-forming oligonucleotides which bind as third strands to duplex DNA in a sequence specific manner. Triplex-forming oligonucleotides may be comprised of any possible combination of nucleotides and modified nucleotides. Modified nucleotides may contain chemical modifications of the heterocyclic base, sugar moiety or phosphate moiety. A high affinity oligonucleotide (Kd≦2×10−8) which forms a triple strand with a specific DNA segment of a target gene DNA is generated. It is preferable that the Kd for the high affinity oligonucleotide is below 2×10−10. The nucleotide binds or hybridizes to a target sequence within a target gene or target region of a chromosome, forming a triplex region. The binding of the oligonucleotide to the target region stimulates mutations within or adjacent to the target region using cellular DNA synthesis, recombination, and repair mechanisms. The mutation generated activates, inactivates, or alters the activity and function of the target gene.
Owner:CONCORD +2
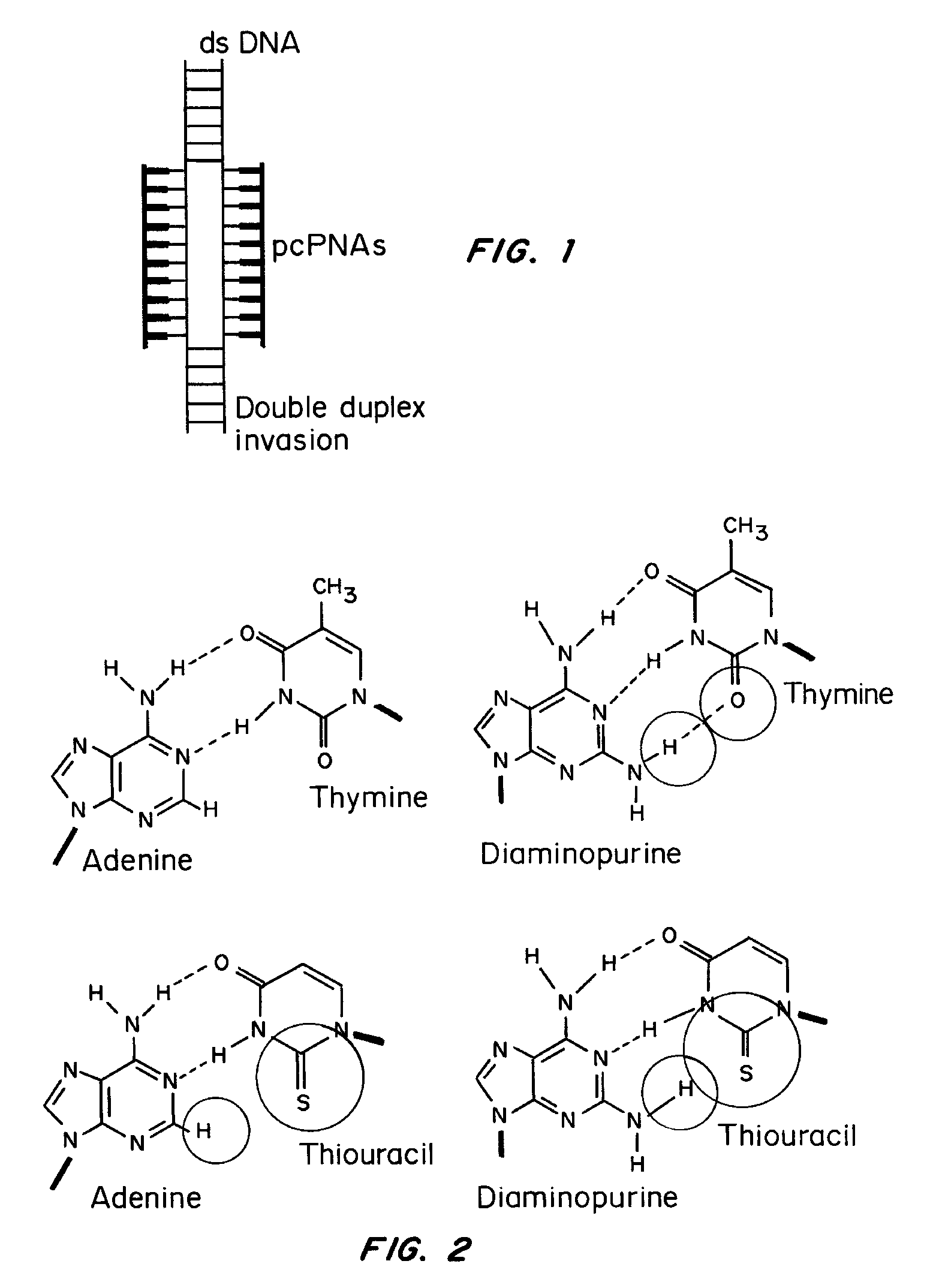
Pseudocomplementary oligonucleotides for targeted gene therapy
ActiveUS8309356B2Increase flexibilityImprove efficiencySugar derivativesCarbohydrate active ingredientsCellular mechanismDouble stranded
Owner:YALE UNIV
Nanofluidic sorting system for gene synthesis and PCR reaction products
ActiveUS20150283514A1Reduce yieldIncrease the lengthSludge treatmentVolume/mass flow measurementPolymerase LGene synthesis
Owner:MASSACHUSETTS INST OF TECH
Plant drug for treatment of liver disease
The present invention related to safe plant drug for treatment of liver disease, specifically, this invention proves a safe plant drug Schisandrin and its preparation. Schisandrin has the following pharmaceutical functions: increasing tumor suppresson genes express activity, decreasing activity of oncogenes, increasing immune function, increasing liver DNA synthesis, decreasing serum alamine aminotransferase activity, increasing glutathione level, increasing glutathione reductase activity, decreasing lipid peroxidation of liver, increasing hepatic microsomal monooxygenases activity, increasing ATP content in liver, increasing energy metabolism activity, decreasing density lipoprotein oxidation, protecting gastrointestinal function, increasing killer cell activity, increasing complement activity, decreasing induced liver cancer activity and decreasing grown of cancer cells.
Owner:ZHAO XINXIAN
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com