Method for predicting membrane protein beta-barrel transmembrane area based on sparse coding and chain training
A sparse coding, transmembrane region technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve problems such as reducing prediction accuracy, and achieve the effects of improving prediction accuracy, avoiding erroneous judgments, and reducing burrs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
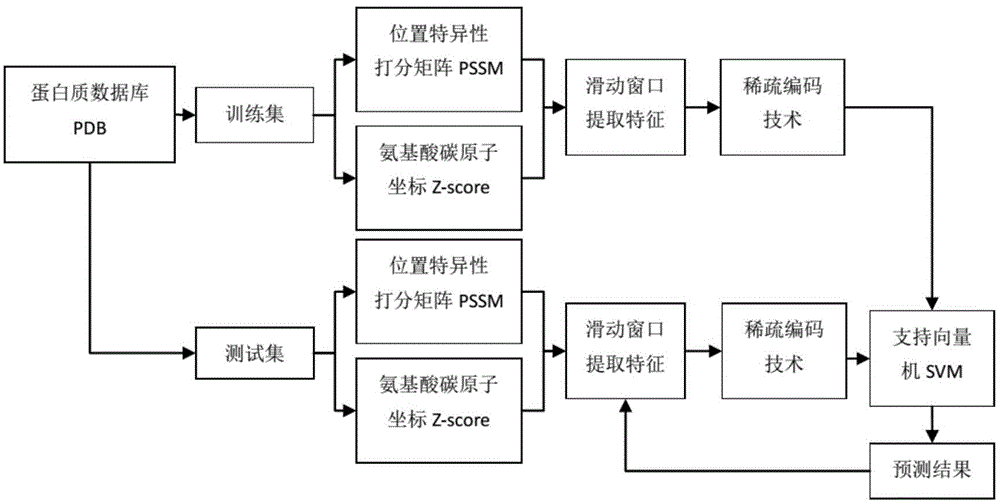
[0025] Such as figure 1 As shown, this embodiment includes the following steps:
[0026] 1) After obtaining the position-specific scoring matrix containing evolution information and the Z coordinate value representing amino acid distance information as features, the position-specific scoring matrix (position-specific scoring matrix, PSSM) generated by the PSI-BALST sequence alignment tool will be and the residue distance information Z-score calculated by the Z-pred software were normalized, in which: the PSSM matrix represents the amino acid sequence evolution information, reflecting the conservation of amino acids, which has been proven to be an effective protein feature; and the Z- socre represents the amino acid distance information to calculate the distance of each residue relative to the membrane center, which is also used as an effective protein characteristic for the studied transmembrane structure.
[0027] The normalization formula for eigenvectors is: f...
Embodiment 2
[0051] This embodiment includes the following steps:
[0052] [1] Protein data set: select the data set from the protein database, and divide the training set and test set respectively;
[0053] [2] Feature selection: respectively select the position-specific scoring matrix PSSM representing protein evolution information and Z-score representing the distance information of amino acids relative to the membrane;
[0054] [3] Feature extraction: extract feature vectors through normalization and sliding window methods;
[0055] [4] Sparse coding: the extracted features are calculated by the method of sparse coding, and the calculated sparse coefficients and base vectors are used to represent the original data. After the experimental statistics, the base vectors and the number of iterations are selected to be 128 and 1000 times respectively;
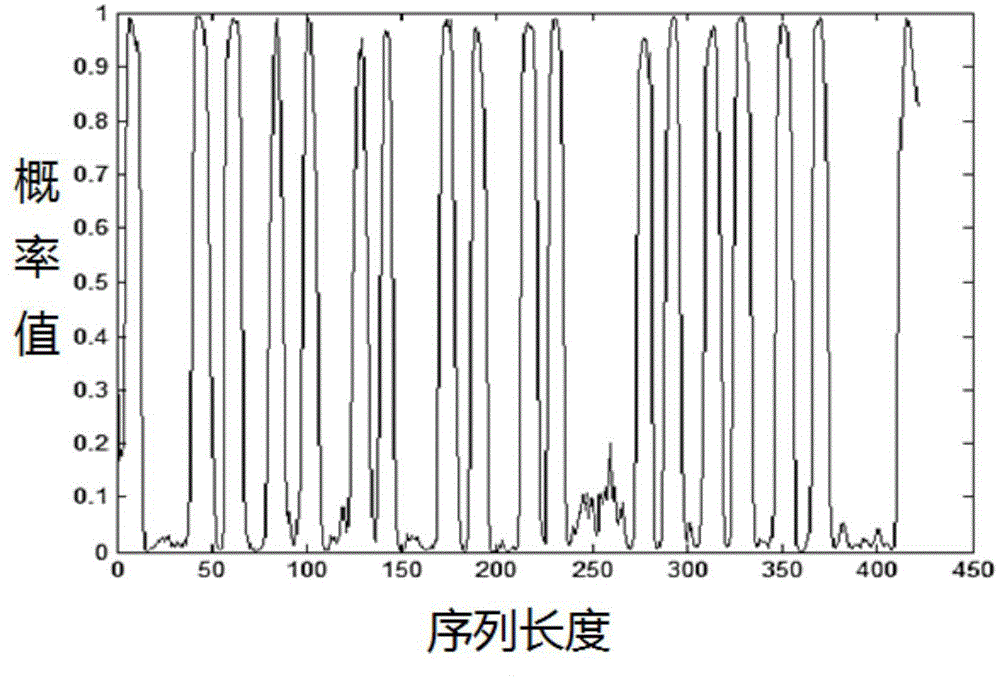
[0056] Support vector machine SVM[5] determines the optimal parameters c and g through 5-fold cross-validation and grid search method, sele...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com