A system for high-throughput testing str typing and its detection method
A high-throughput and systematic technology, applied in the biological field, can solve the problems of limiting the number of detectable loci, unable to measure STR sequence polymorphism, unable to quickly detect DNA information, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
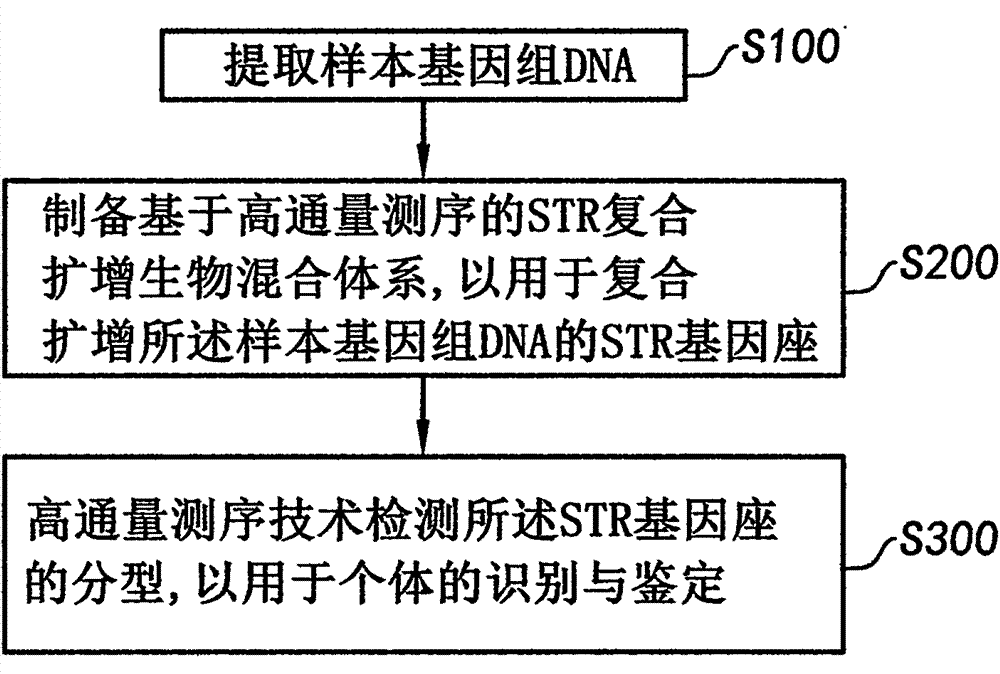
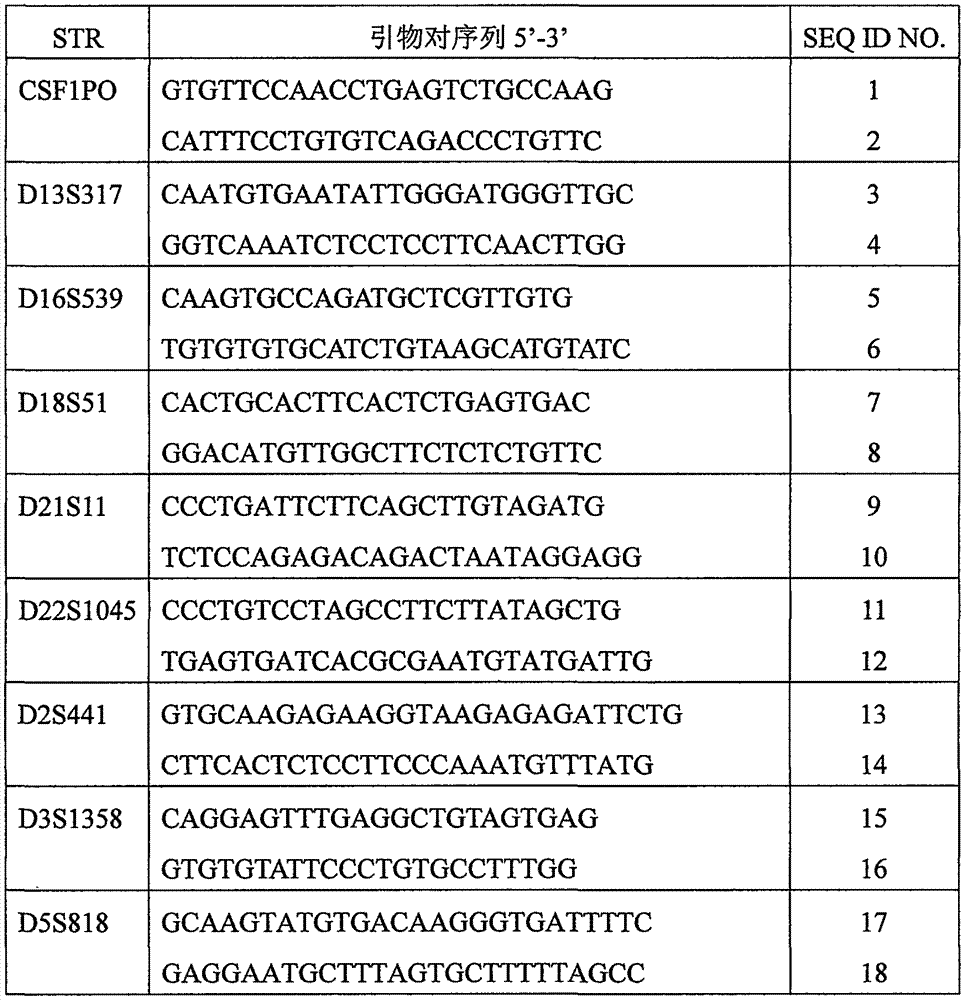
[0031]A system for high-throughput testing of STR typing includes a biological mixture system comprising at least one sample solution and at least one different primer pair for an STR locus, wherein the primer pair can be used to amplify Increase at least one of the STR loci, the STR is selected from autosomes and / or Y chromosomes; a multiplex amplification system, the multiplex amplification system is used for multiplex amplification of the STR by the at least one different primer pair Gene loci, forming at least one STR locus amplification product; and a detection system, wherein the detection system is a high-throughput sequencing device for detecting at least one type of the STR loci. Therefore, the autosome and / or Y chromosome STR compound amplification typing detection can be realized through high-throughput sequencing technology, and the detection accuracy and the recognition degree of individual identification can be improved.
[0032] Wherein, when the STR is selected...
Embodiment 2
[0117] A system for high-throughput testing of STR typing includes a biological mixture system comprising at least one sample solution and at least one different primer pair for an STR locus, wherein the primer pair can be used to amplify Increase at least one of the STR loci, the STR is selected from autosomes and / or Y chromosomes; a multiplex amplification system, the multiplex amplification system is used for multiplex amplification of the STR by the at least one different primer pair Gene loci, forming at least one STR locus amplification product; and a detection system, wherein the detection system is a high-throughput sequencing device for detecting at least one type of the STR loci. Therefore, the rapid detection of STR typing on autosomes and / or Y chromosomes can be realized by high-throughput sequencing technology, and the detection accuracy and the recognition degree of individual identification can be improved.
[0118] Wherein, when the STR is selected from the Y c...
Embodiment 3
[0177] A system for high-throughput testing of STR typing includes a biological mixture system comprising at least one sample solution and at least one different primer pair for an STR locus, wherein the primer pair can be used to amplify Increase at least one of the STR loci, the STR is selected from autosomes and / or Y chromosomes; a multiplex amplification system, the multiplex amplification system is used for multiplex amplification of the STR by the at least one different primer pair Gene loci, forming at least one STR locus amplification product; and a detection system, wherein the detection system is a high-throughput sequencing device for detecting at least one type of the STR loci. Therefore, the autosome and / or Y chromosome STR compound amplification typing detection can be realized through high-throughput sequencing technology, and the detection accuracy and the recognition degree of individual identification can be improved.
[0178] Wherein, when the STR is selecte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com