Preparation method of tumor sample sequencing reference
A reference product and sequencing technology, applied in the field of tumor sample sequencing reference product preparation, can solve the problems of high diagnostic error rate, lack of high quality, scientific and systematic comparison of results, etc., to achieve clear genetic background, good repeatability, and ensure standardization sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
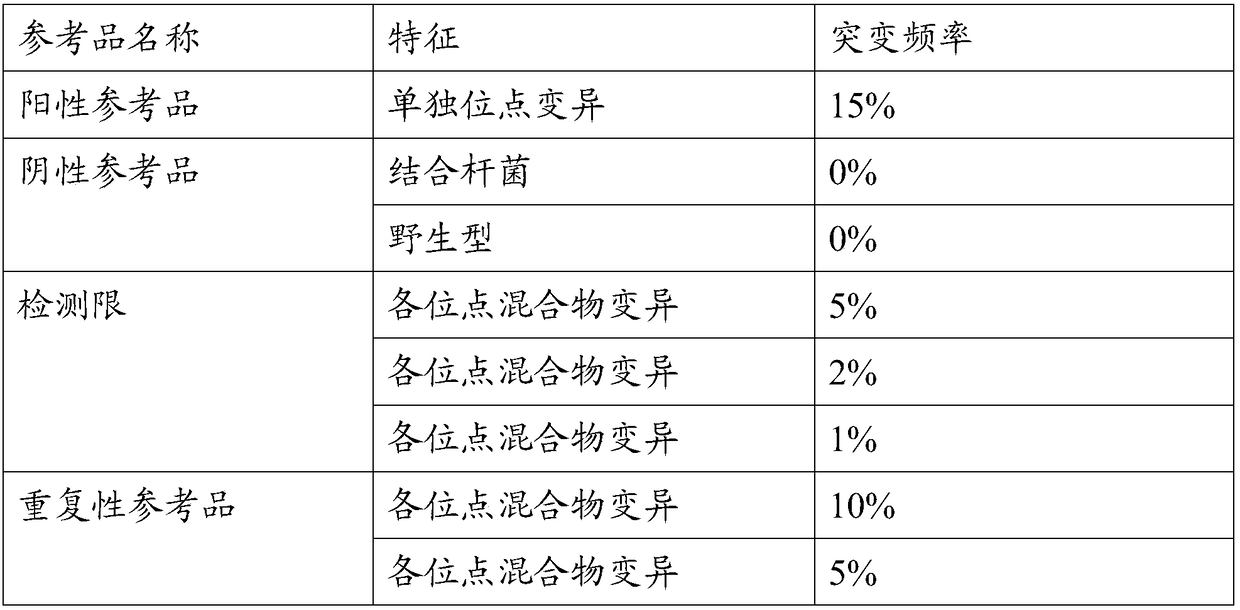
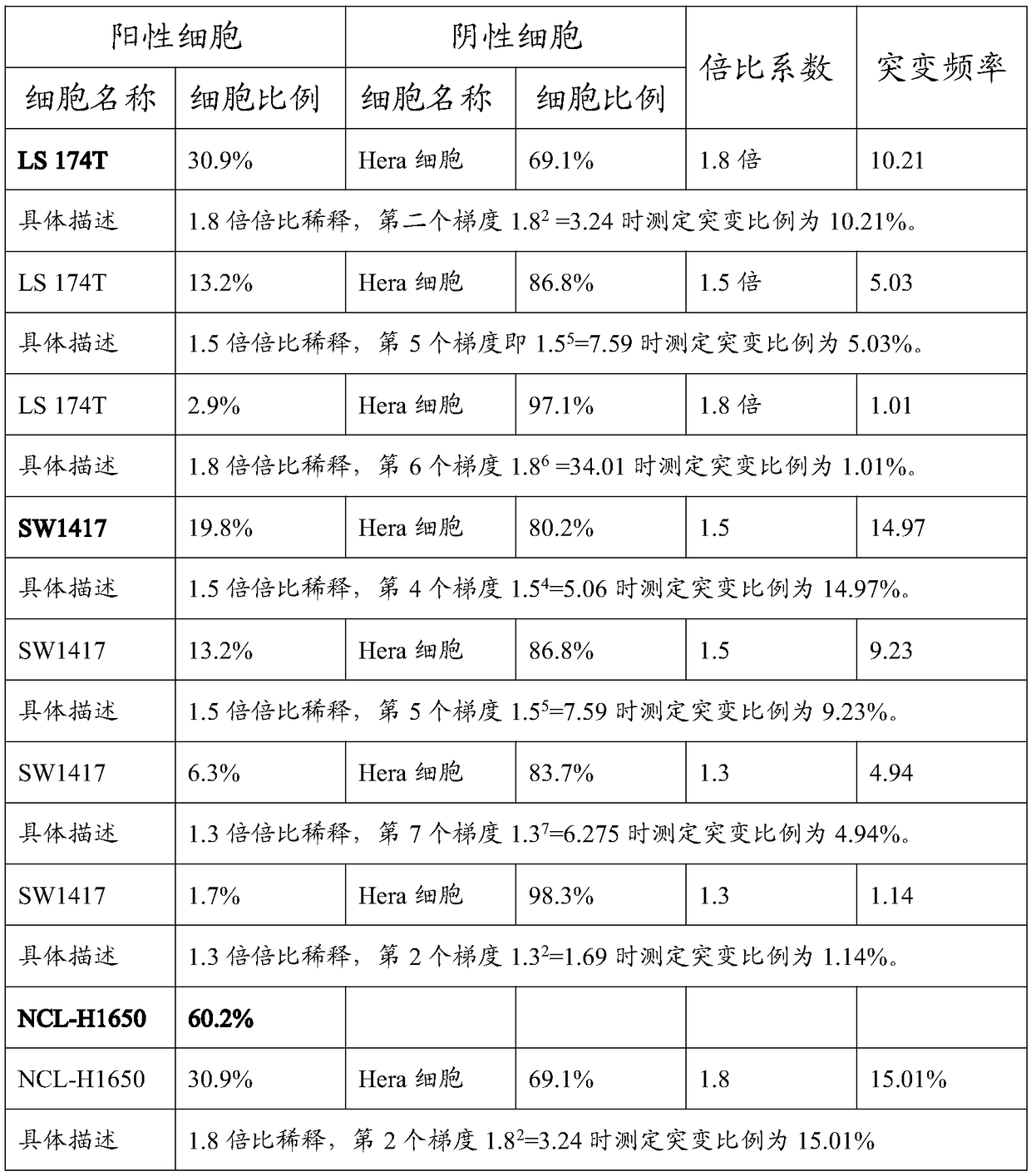
[0021] The invention provides a method for preparing a tumor sample sequencing reference product, which comprises the following steps: 1) performing monoclonal culture on tumor cell lines to obtain a monoclonal cell line; 2) extracting the whole genome of the monoclonal cell line; 3) using Perform high-throughput sequencing on the whole genome of the monoclonal cell line to determine the allele frequency to obtain the allele mutation frequency; 4) determine the dilution according to the allele mutation frequency obtained in step 3) and the target allele mutation frequency multiple; 5) after the monoclonal cell line was digested with trypsin, the cell concentration of the monoclonal cell line was obtained by counting with a flow cytometer, in terms of cell number, and the corresponding number of allele mutation frequency was 0 negative cell times Ratio diluting the monoclonal cell line to obtain mixed cells; 6) extracting the whole genome of the mixed cells in step 5), and perfo...
Embodiment 1
[0033] A new method for producing lung cancer high-throughput sequencing reference products using cell lines, comprising the following steps:
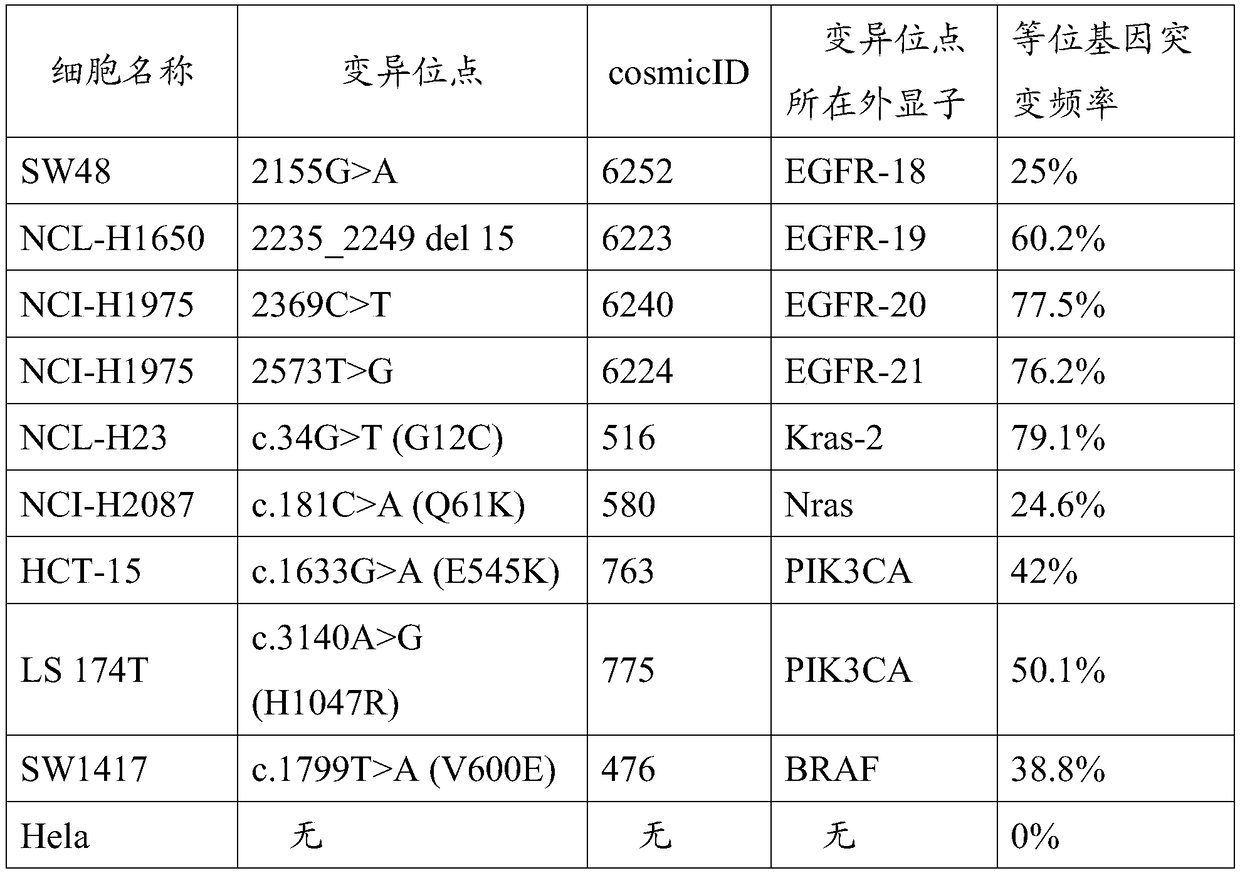
[0034] 1) Perform monoclonal culture screening of cell lines purchased from the American Type Culture Collection (ATCC), involving 8 cell lines as follows: NCI-H1650, NCI-H1975, NCI-H23, NCI-H2087, HCT-15 , LS 174T, SW1417, SW48; The monoclonal screening of cell lines adopts a dilution culture scheme, as follows: After counting cells, they are divided into 96-well plates, and the number of cells in each well is guaranteed to be 0.5, which makes single cells appear in some wells , carry out cell culture amplification on these; the final standard for determining monoclonal is to extract the genome from the cultured and expanded cells as a template, carry out high-throughput library construction, and test its allele frequency. Those with stable frequency after multiple tests are Screen qualified cell lines.
[0035] 2) The entire genome ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com