Molecular marker identification method related to pig birth litter weight and fetal weight and application of molecular marker identification method
A technology of molecular markers and identification methods, applied in the fields of biochemical equipment and methods, microbial determination/inspection, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] A molecular marker identification method related to pig birth weight and fetal weight:
[0093] (1) Sample collection: put in 75% alcohol and store at -20°C, DNA to be extracted;
[0094] (2) Extraction and detection of pig genomic DNA:
[0095] 1) Genomic total DNA was extracted from pig tissue;
[0096] 2) After the DNA is fully dissolved, measure the concentration of the extracted DNA with a UV spectrophotometer, and detect the quality of the extracted DNA by agarose gel electrophoresis;
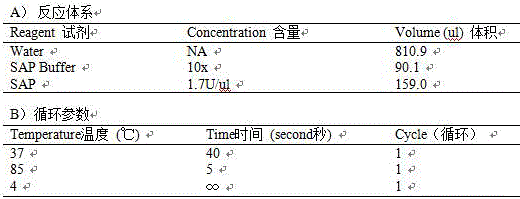
[0097] (3) SNP chip genotype determination and quality control of genotype data:
[0098] 1) Take 2ul stock solution for electrophoresis detection
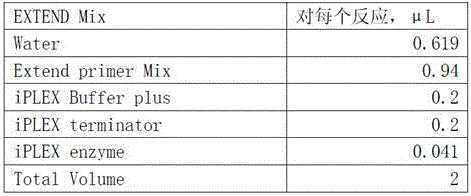
[0099] 2) Primer design: According to the SNP site sequence information, use the primer design software Assaydesign3.1 of sequenom company to design and synthesize the PCR reaction and single-base extension primers;
[0100]
[0101] Take 2.5ul each of Forward PCR primer and Rewerse PCR primer for each SNP, and mix them into a 5...
Embodiment 2
[0113] A molecular marker identification method related to pig birth weight and fetal weight:
[0114] 1. Primer design:
[0115] 1st-PCRP: forward primer (F)
[0116] 2nd-PCRP: reverse primer (R)
[0117] UEP_SEQ: Single base extension primer
[0118] UEP_MASS: single base extension primer quality
[0119] UEP_DIR: the DNA strand where the single base extension primer is located
[0120] F: chain of justice
[0121] R: antisense strand
[0122] 2. Experimental process:
[0123] a. First dilute the primer well, primer dilution:
[0124] (1) For each SNP, there are three primers, and the numbers of these three primers are marked on the cap of the primer synthesis tube.
[0125] (2) Forward PCR primers and Rewerse PCR primers are first centrifuged and then added with water. The amount of water added is related to the OD value (the primer tube and the primer design table are marked), and 36ul of water is added for every 1OD. After adding water, let stand at room temperat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com