Efficient high-accuracy whole-genome selection method capable of performing parallel operation
A genome-wide and accurate technology, applied in genomics, proteomics, instruments, etc., can solve the problems of complex parameter solving process, poor accuracy of breeding values, complex assumptions, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0036] The present invention will be further described below in conjunction with the accompanying drawings and specific embodiments.
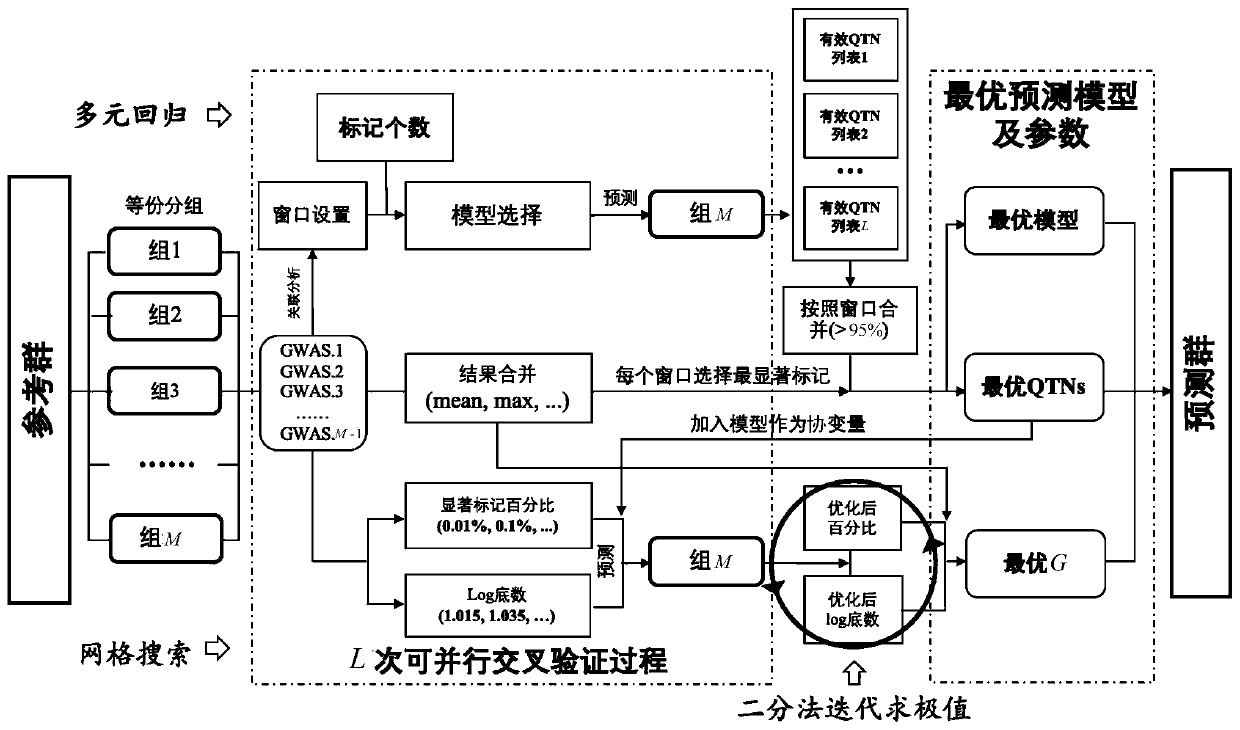
[0037] Such as figure 1 As shown, the high-efficiency parallel computing and high-accuracy genome-wide selection method of the present invention is characterized in that it comprises the following steps:
[0038] Step 1: read the original genotype file and the original phenotype file, extract the genotype data and phenotype data of the same individual in the original genotype file and the original phenotype file, and form a new genotype file and a new phenotype file, And use the new genotype file to calculate the kinship matrix G among all individuals.
[0039] In this embodiment, the S4 data format in the R language is used to establish the data mapping between the disk and the internal memory; the reading and storage of the genotype file and the phenotype file adopt the big.matrix format in the R CRAN::bigmemory software package , and provi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com