Early detection and prognosis of colon cancers
A technology for colorectal cancer, colon, in the field of aberrant methylation patterns
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] Materials and Method
[0097] [88] Cell culture and processing. HCT116 cells and isogenic genetic knockout derivatives are maintained as previously described (14). For drug treatment, logarithmic phase CRC cells were cultured in McCoys 5A medium (Invitrogen) for 96 hours. McCoys 5A medium contained 10% BCS and 1× penicillin / streptomycin and 5 μM 5 aza-deoxycytidine (DAC ) (Sigma; stock solution: 1 mM in PBS), changing the medium and DAC every 24 hours. The cells were treated with 300 nM Trichostatin A (Sigma; stock solution: 1.5 mM dissolved in ethanol) for 18 hours. The control cells were simulated in parallel, and an equal volume of PBS or ethanol without drugs was added.
[0098] [89] Microarray analysis. Following the manufacturer's instructions, including DNase digestion steps, use Triazol (Invitrogen) and RNeasy kit (Qiagen) to harvest total RNA from log phase cells. NanoDropND-100 was used, followed by 2100 Bioanalyzer (Agilent Technologies) for quality evaluatio...
Embodiment 2
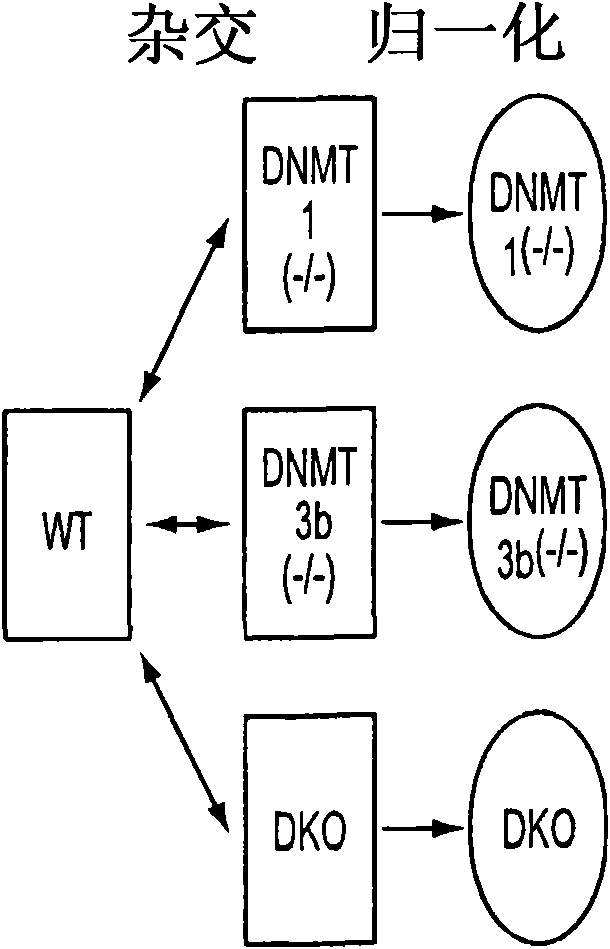
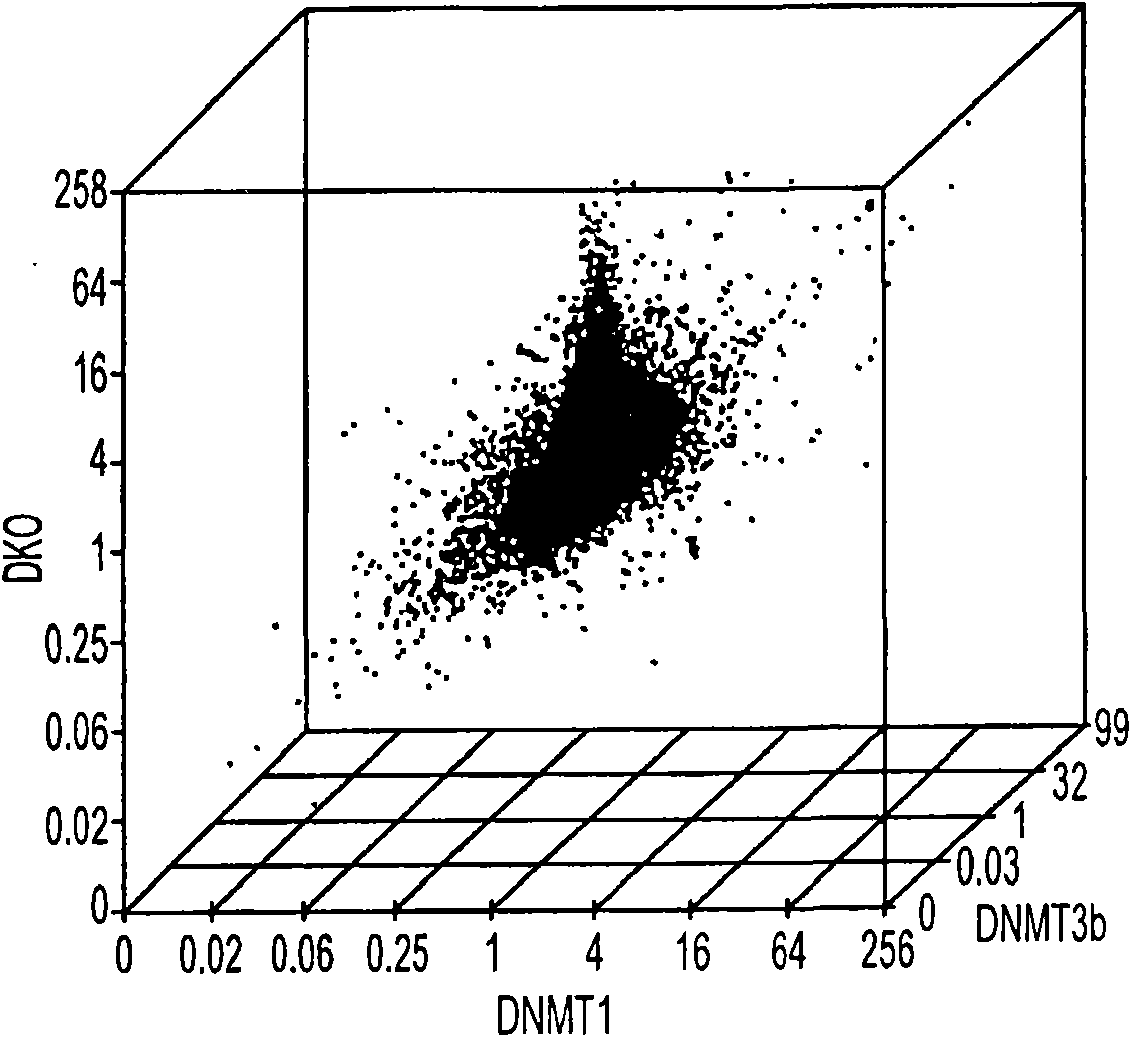
[0103] [93] For the comprehensive identification of hypermethylation-dependent gene expression changes, our first step is to compare wild-type HCT116CRC cells with a single cell carrying two major human DNA methyltransferases in a genome-wide expression array-based method. And the isogenic chaperone cell of the combined gene deletion ( Figure 1A , 14). Importantly, in DNMT1 (- / -) DNMT3b (- / -) Double knockout (DKO) HCT116 cells-which actually completely lost all 5-methylcytosine, all previously single-checked hypermethylated genes-lacking basic expression in wild-type cells-suffered Promoters accompanying gene re-expression are demethylated (14-17). By grading genes according to the altered signal intensity on the 44K Agilent Technologies array platform, when compared with isogenic wild-type parent cells or isogenic cell lines, we observed a unique peak of increased gene expression in DKO cells. Gene wild-type parent cell or isogenic cell line 1 or 3b of DNMT has been deleted ...
Embodiment 3
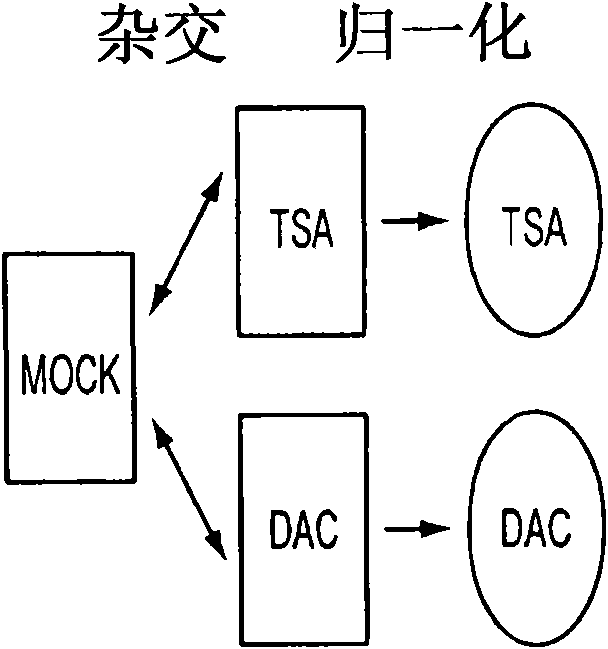
[0106] [95] To illustrate the sensitivity of our new array method to identify CpG island hypermethylated genes, we first examined 11 genes known to be hypermethylated, completely silenced, and re-expressed after DAC treatment in HCT116 cells (Figure 4(S1A))(14-17). All tested genes remained in the TSA unreacted zone (Figure 4 (S1B and C)), and the direction of expression change was fully correlated in DAC treatment and DKO cells (Figure 4 (S1D)). Importantly, for the increase in DAC, 5 guide genes (45%) increased by a factor of 2 or more, and another 3 genes or a total of 73% of genes increased by a factor of 1.3 or more.
[0107] [96] Based on the observed sensitivity difference between DKO and DAC-induced gene increase and the behavior of the guide gene in the array platform, in the TSA negative region, we specified the upper layer of the gene (two-fold increase or more) and The lower layer (increase between 1.4-fold and 2-fold) to identify hypermethylated oncogenes ( Figure ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com