ARMS fluorescent quantitative PCR-based gene mutation kit and method thereof
A fluorescence quantitative and kit technology, applied in the field of molecular biology, can solve the problems of unfavorable result judgment and high background, and achieve the effects of accurate and reliable result judgment, reduced pollution, and fast and convenient detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
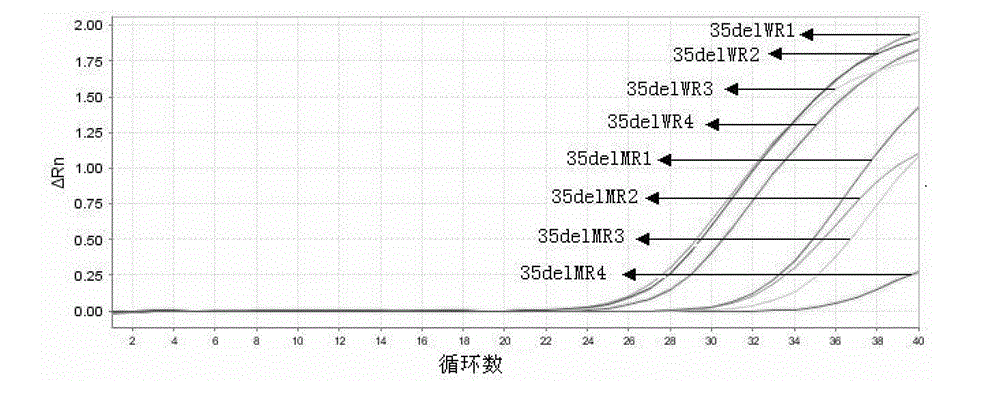
[0050] Example 1 Detection of GJB235delG deletion mutations associated with deafness using conventional ARMS primers and enhanced ARMS primers of the present invention
[0051] In the present invention, on the basis of conventional ARMS primers, a mismatched base is set in the primer at the 6th to 8th upstream base of the deletion site in the direction of extension, thereby reducing the number of wild-type bases in real-time fluorescent quantitative PCR. The amplification efficiency of enhanced ARMS primers for mutant samples and the amplification efficiency of mutant enhanced ARMS primers for wild-type samples make the results easier to judge.
[0052] 1) Design of ARMS primers and enhanced ARMS primers
[0053] Three pairs of wild-type ARMS primers, three pairs of mutant ARMS primers, one pair of wild-type enhanced ARMS primers and one pair of mutant enhanced ARMS primers were designed in the experiment. 60.0°C, GC value 40.0%-60.0%, primer size 20±3bp. According to the sp...
Embodiment 2
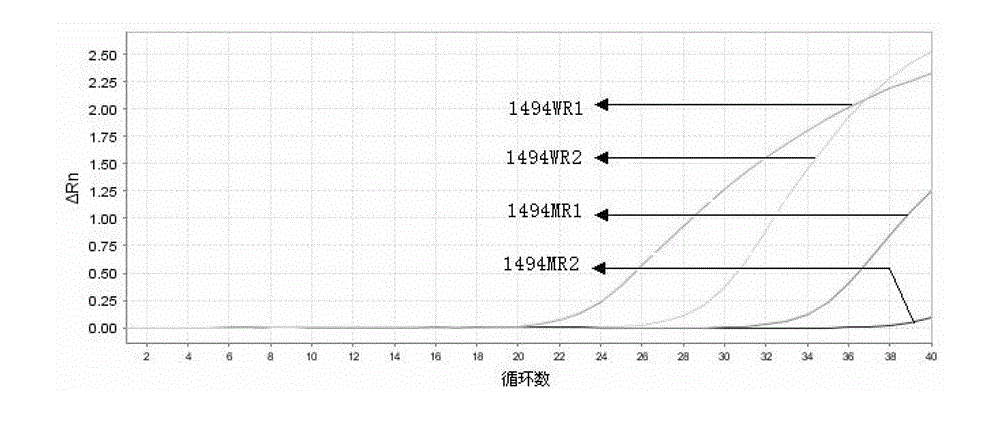
[0072] Example 2 Detection of deafness-related gene 12SrRNA1494C>T point mutation using conventional ARMS primers and enhanced ARMS primers of the present invention
[0073] 1) Design of ARMS primers and enhanced ARMS primers
[0074]One pair of wild-type ARMS primers, one pair of mutant ARMS primers, one pair of wild-type enhanced ARMS primers and one pair of mutant enhanced ARMS primers were designed in the experiment. The common upstream primer was 12SrRNAF. 60.0°C, GC value 40.0%-60.0%, primer size 20±3bp. The 3' terminal bases of ARMS primers and enhanced ARMS primers are located at the sites to be tested and are matched with wild-type and mutant genes respectively. In order to improve specificity, introduce One mismatched base, and a mismatched base was introduced at the 4th base and the 13th base at the 3' end of the enhanced ARMS primer respectively. The designed ARMS primer and enhanced ARMS primer sequences are as follows:
[0075] Downstream ARMS primers:
[0076...
Embodiment 3
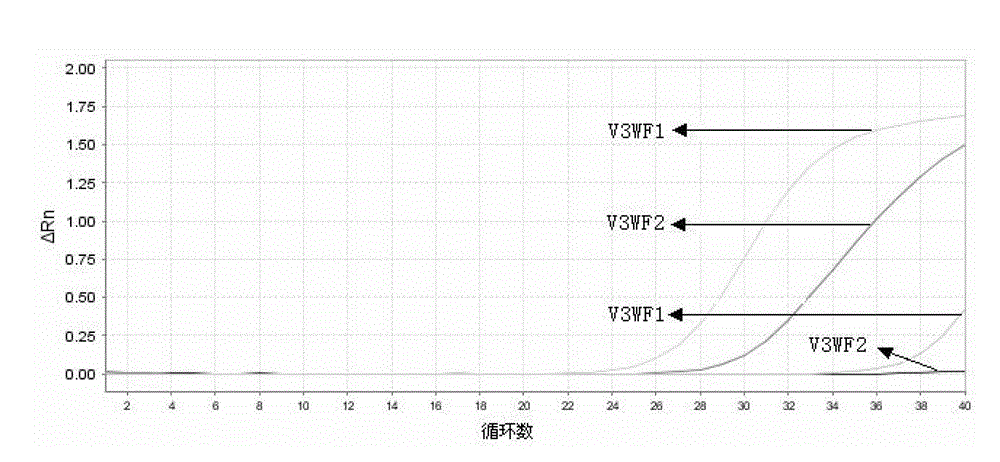
[0090] Example 3 Using conventional ARMS primers and enhanced ARMS primers of the present invention to detect warfarin medication-related sites VKORC13730G>A
[0091] 1) Design of ARMS primers and enhanced ARMS primers
[0092] One pair of wild-type ARMS primers, one pair of mutant-type ARMS primers, one pair of wild-type enhanced ARMS primers and one pair of mutant-type enhanced ARMS primers were designed in the experiment, and the common downstream primer was V3R. 60.0°C, GC value 40.0%-60.0%, primer size 20±3bp. The 3' terminal bases of ARMS primers and enhanced ARMS primers are located at the site to be tested and are matched with wild-type and mutant genes respectively. In order to improve specificity, a Mismatched bases, and a mismatched base was introduced at the 2nd base and the 7th base at the 3′ end of the enhanced ARMS primer. The designed ARMS primer and enhanced ARMS primer sequences are as follows:
[0093] Upstream ARMS primers:
[0094] V3WF1CCCTCCTCCTGCCATA...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com