Patents
Literature
49 results about "Physical Maps" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
What Is a Physical Map? The main objective of a physical map is to show landforms, bodies of water and other geographical features. Features shown include mountains, deserts and lowlands as well as lakes, rivers and oceans. Physical maps show the natural details of the land and water included in the ...
Video surveillance, storage, and alerting system having network management, hierarchical data storage, video tip processing, and vehicle plate analysis
ActiveUS7382244B1Television system detailsFrequency-division multiplex detailsVideo monitoringDriver/operator
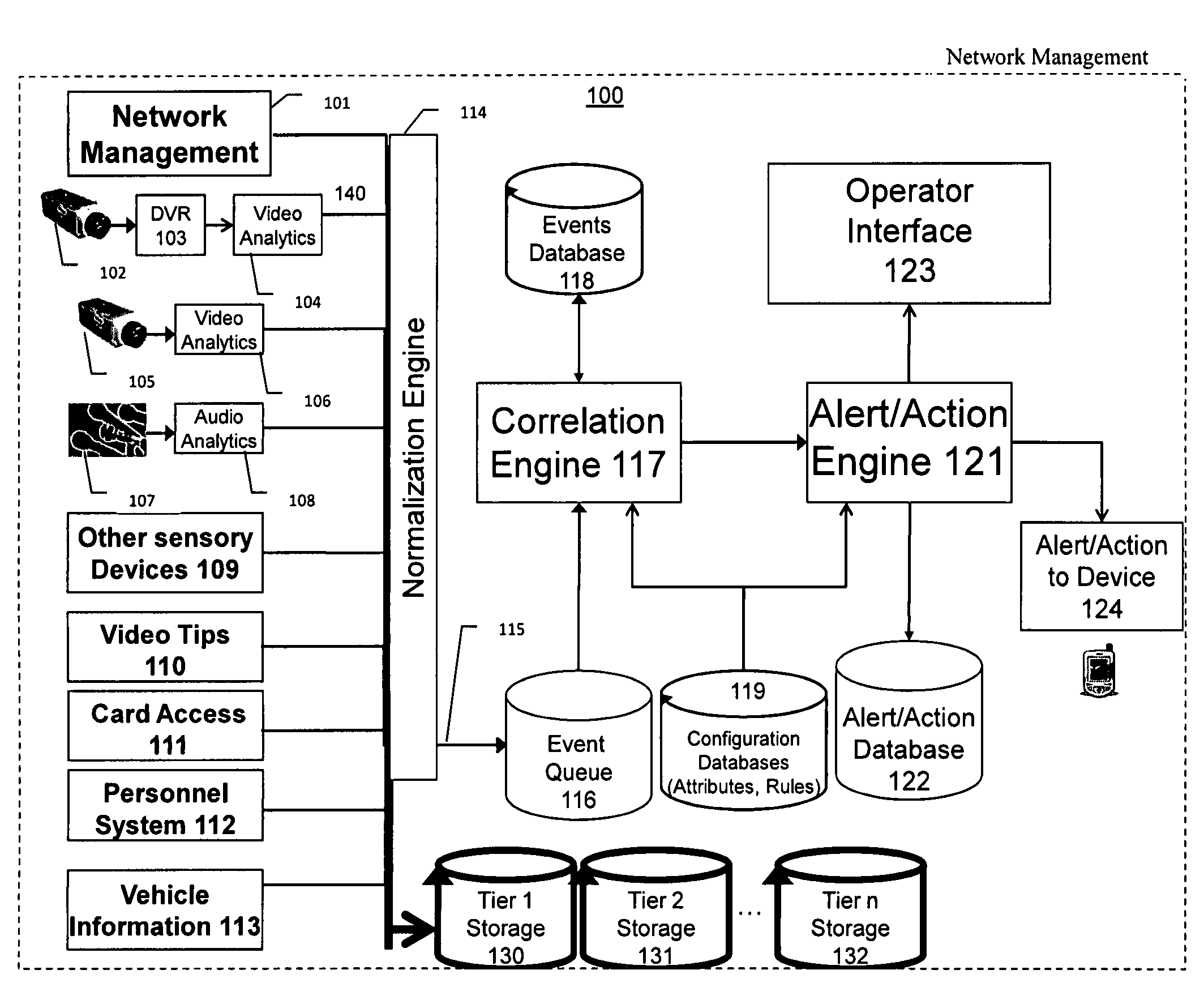
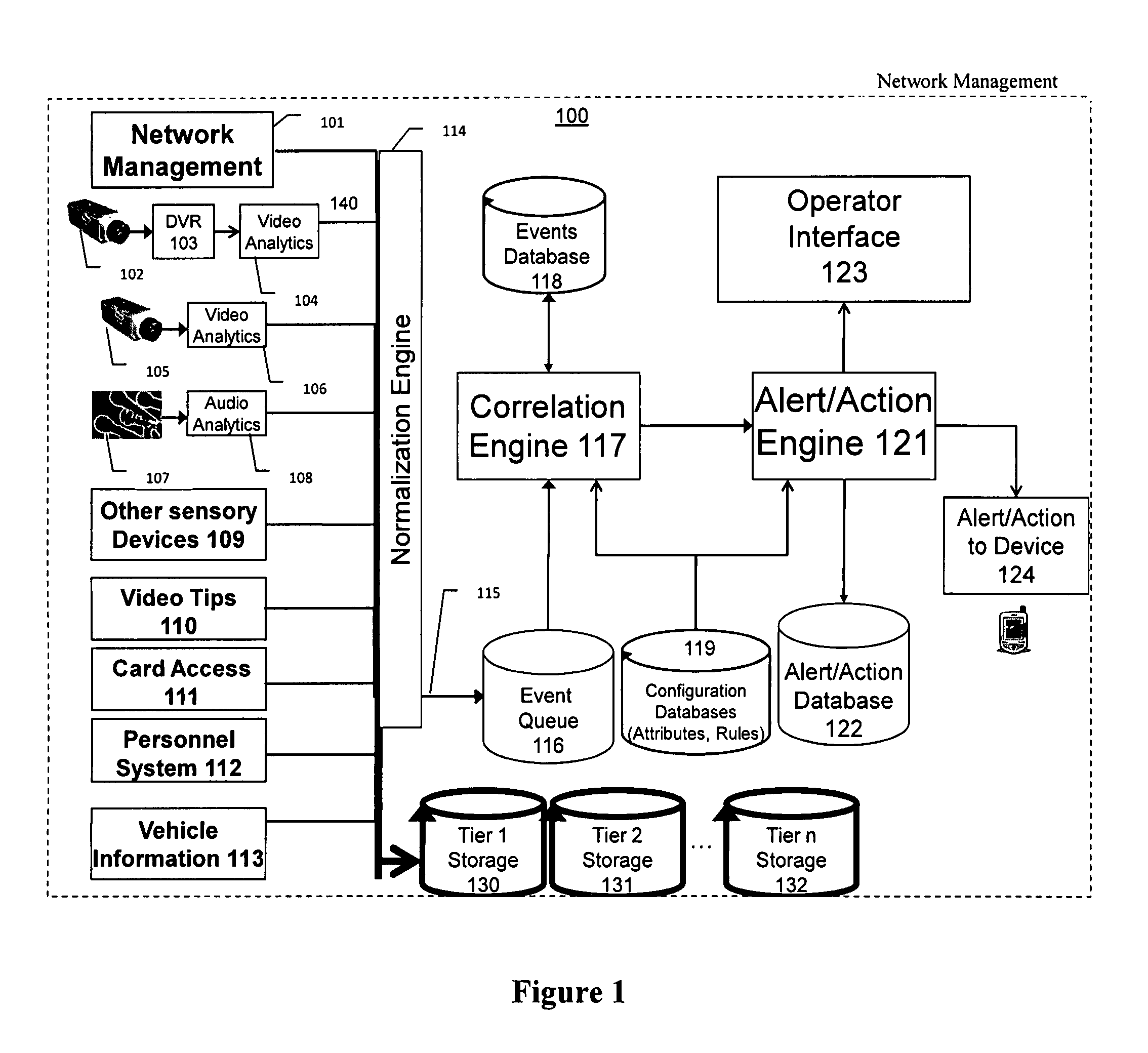
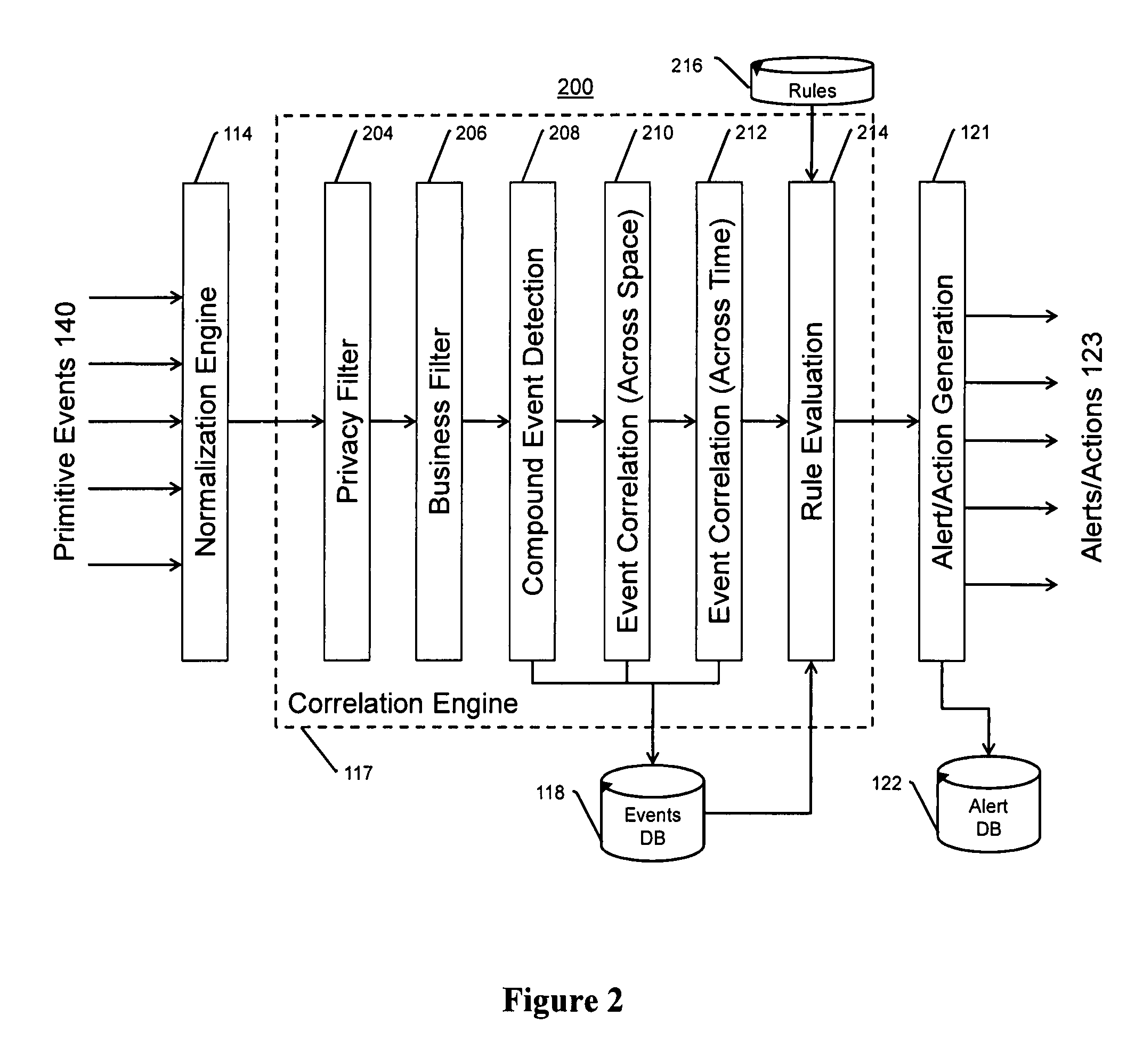
The present invention is a video surveillance, storage, and alerting system having surveillance cameras, video analytics devices, audio sensory devices, other sensory devices, and a plurality of data storage devices. A network management module monitors network status of all subsystems including cameras, servers, storage devices, etc. and shows actively monitored areas on a physical map. A vehicle information module retrieves information from a law enforcement database about vehicles detected in the video data based on the vehicle's license plate, including information about stolen vehicles, as well as warrant, wanted person, and mug shot information for registered drivers of the vehicles. Video tips are received and processed from anonymous and non-anonymous sources. A correlation engine correlates primitive events and compound events from each of the subsystems, weighted by attributes of the events, across both space and time, and an alerting engine generates alerts and performs actions based on the correlation. A hierarchical storage manager manages storage of the vast amounts of data, including video data, based on importance of the data calculated from attributes of the data. A privacy filter ensures no private data is detected, correlated, or stored.
Owner:SECURENET SOLUTIONS GRP
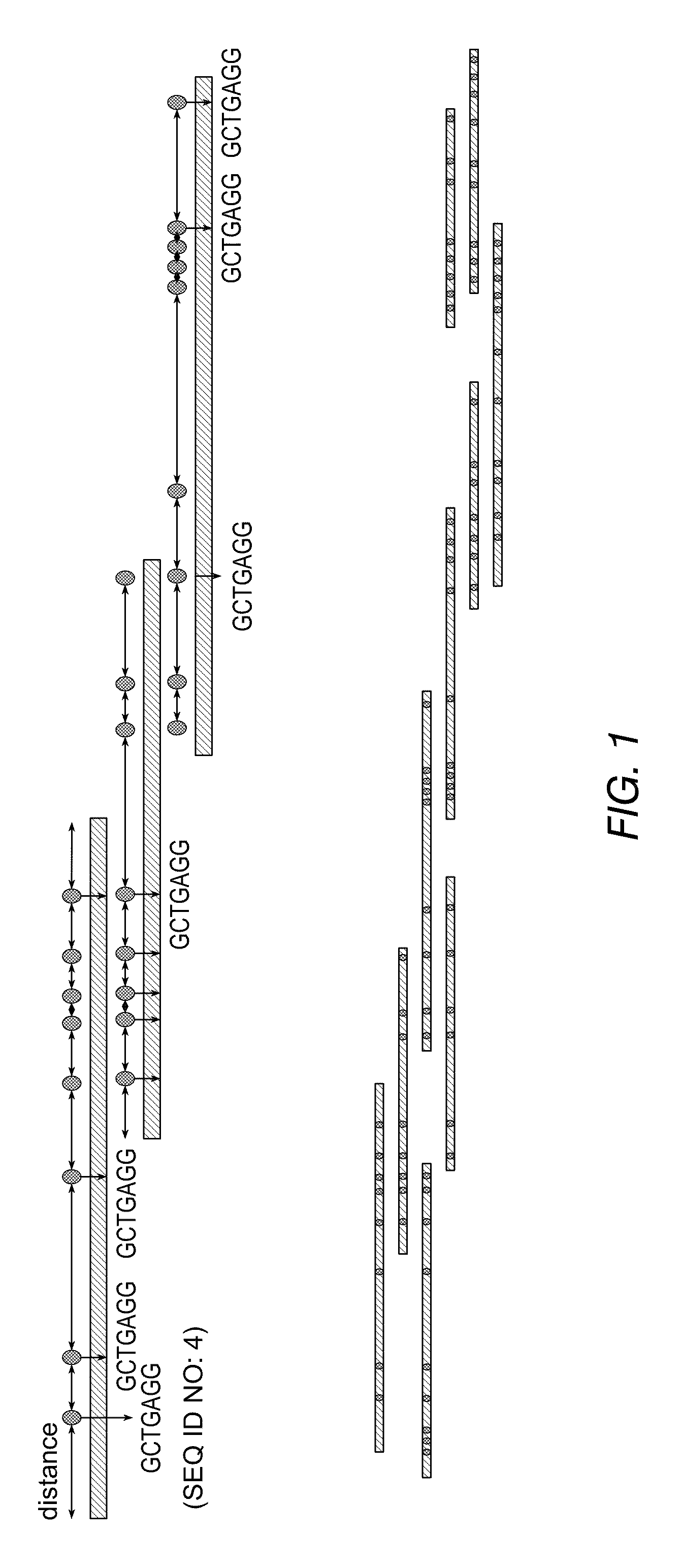
Method and compositions for ordering restriction fragments
InactiveUS7598035B2High density physicalPrecise positioningSugar derivativesMicrobiological testing/measurementPhysical MapsNucleotide sequencing
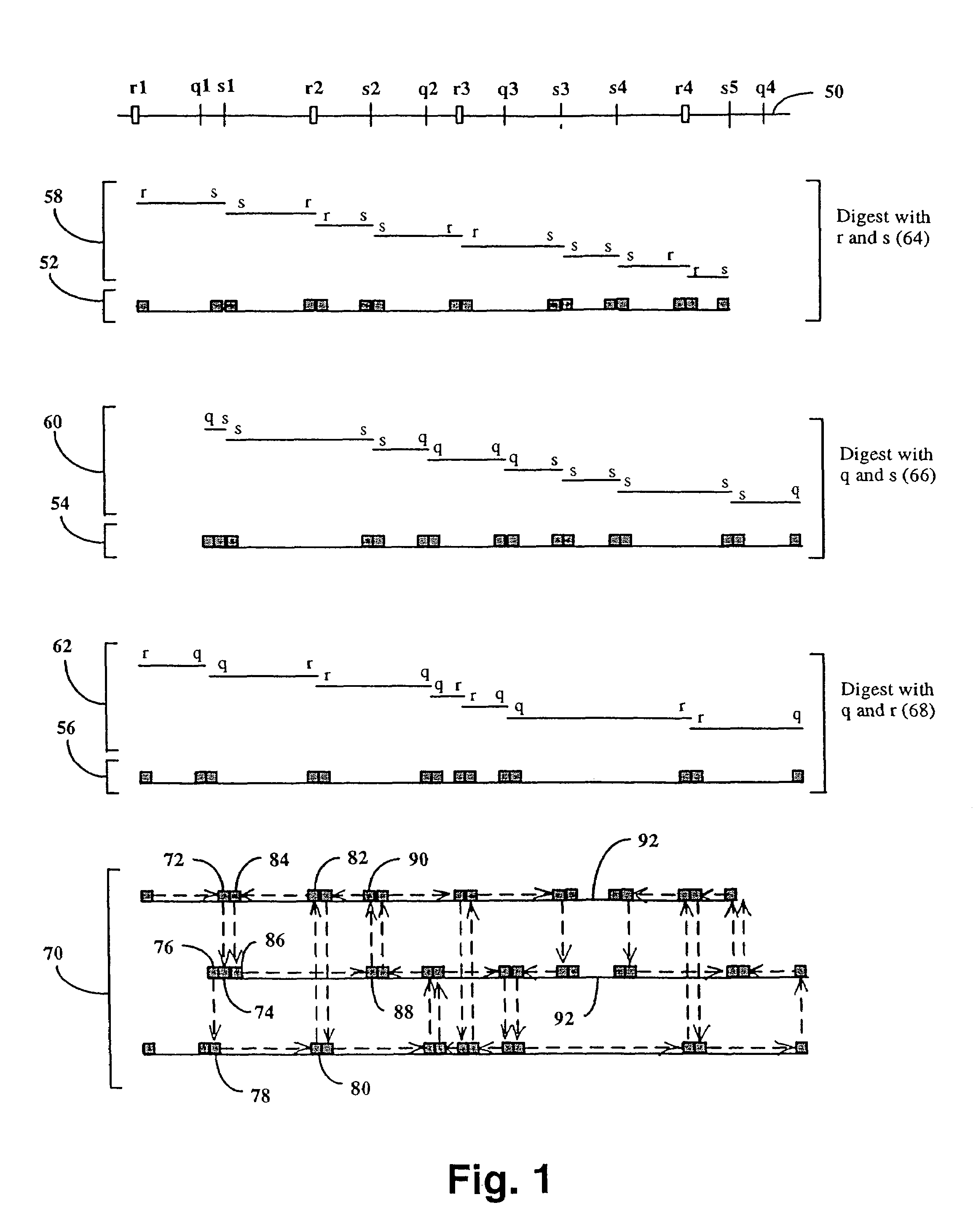
The invention provides a method for constructing a high resolution physical map of a polynucleotide. In accordance with the invention, nucleotide sequences are determined at the ends of restriction fragments produced by a plurality of digestions with a plurality of combinations of restriction endonucleases so that a pair of nucleotide sequences is obtained for each restriction fragment. A physical map of the polynucleotide is constructed by ordering the pairs of sequences by matching the identical sequences among the pairs.
Owner:ILLUMINA INC
Parallel methods for genomic analysis
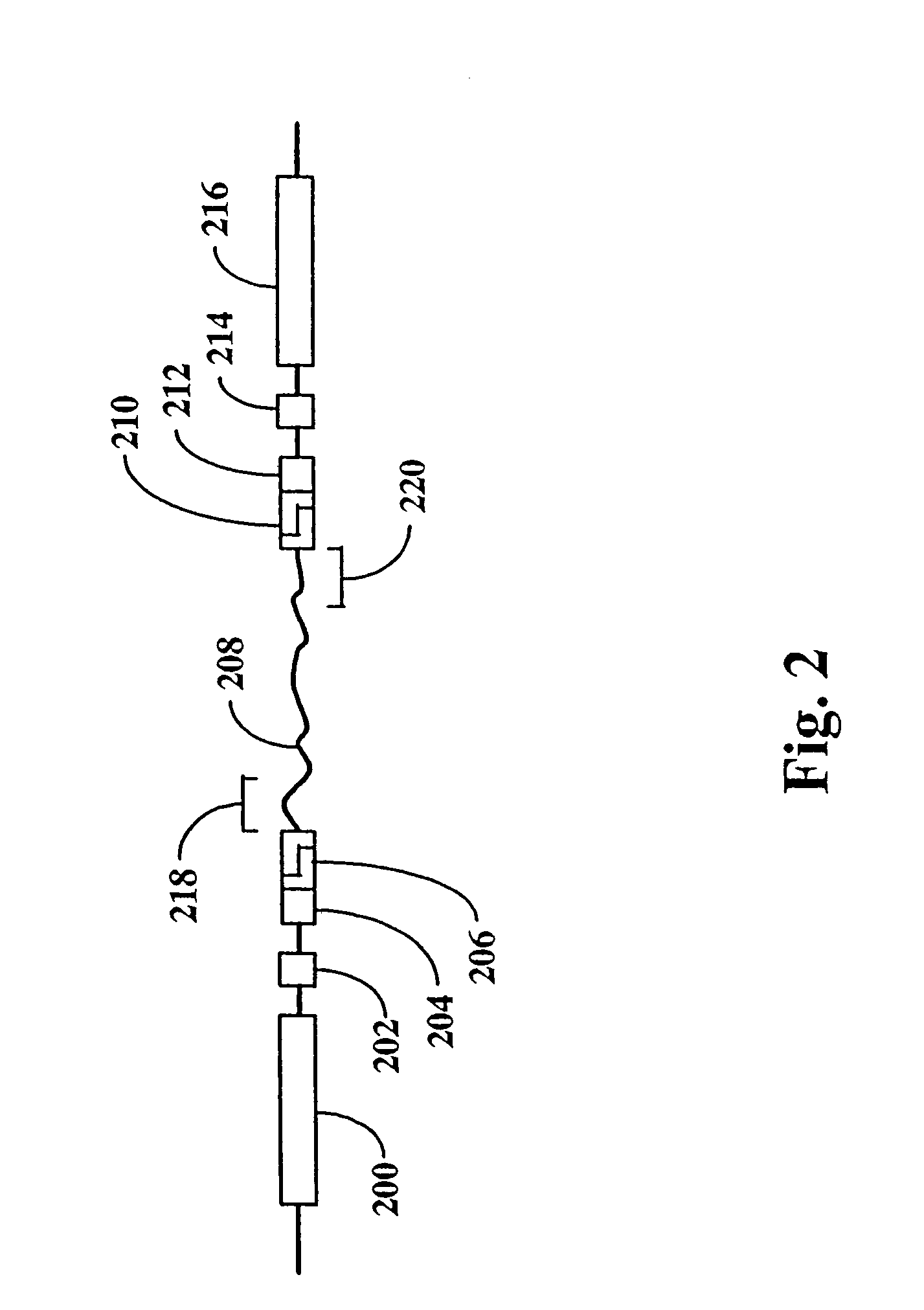
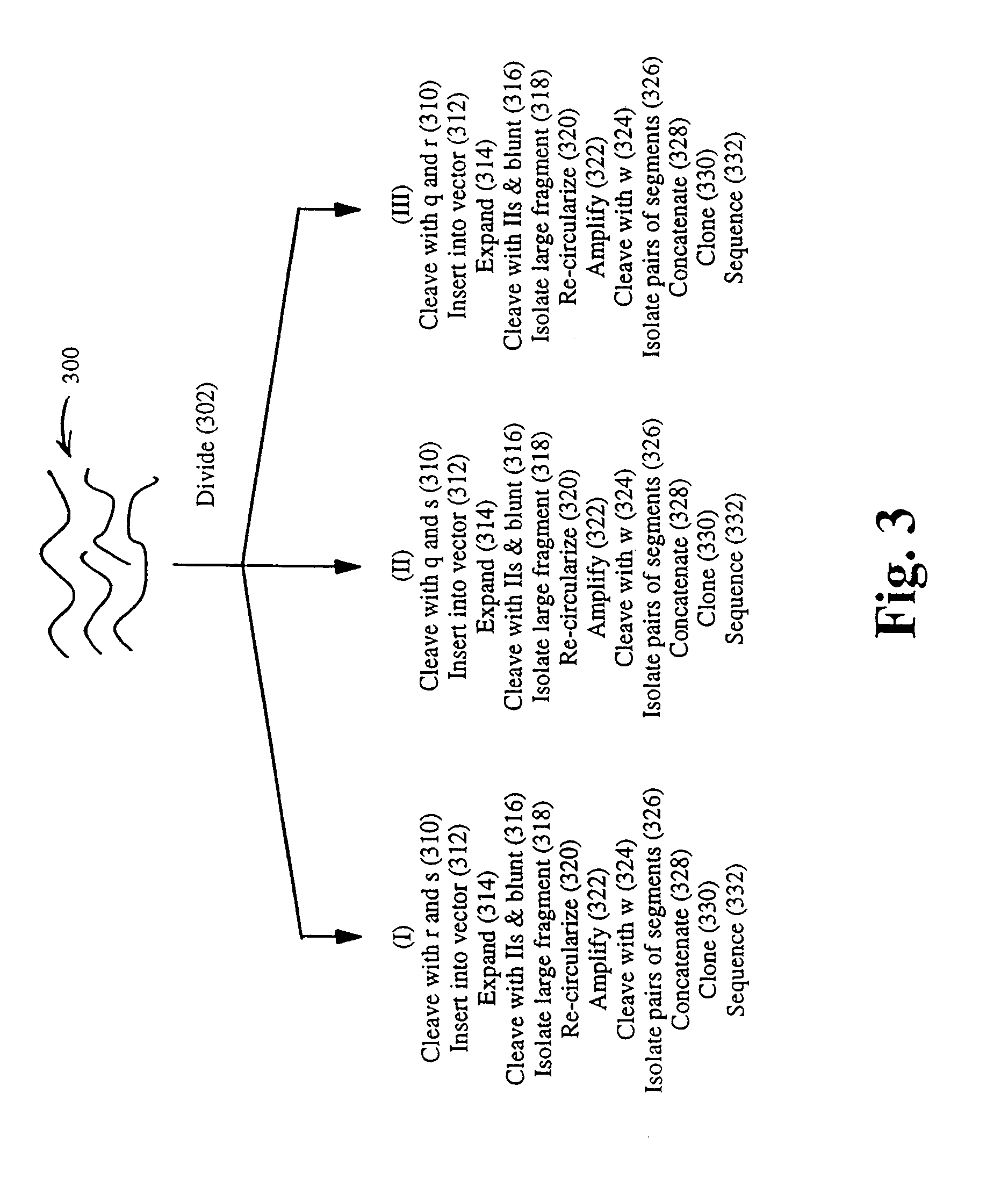
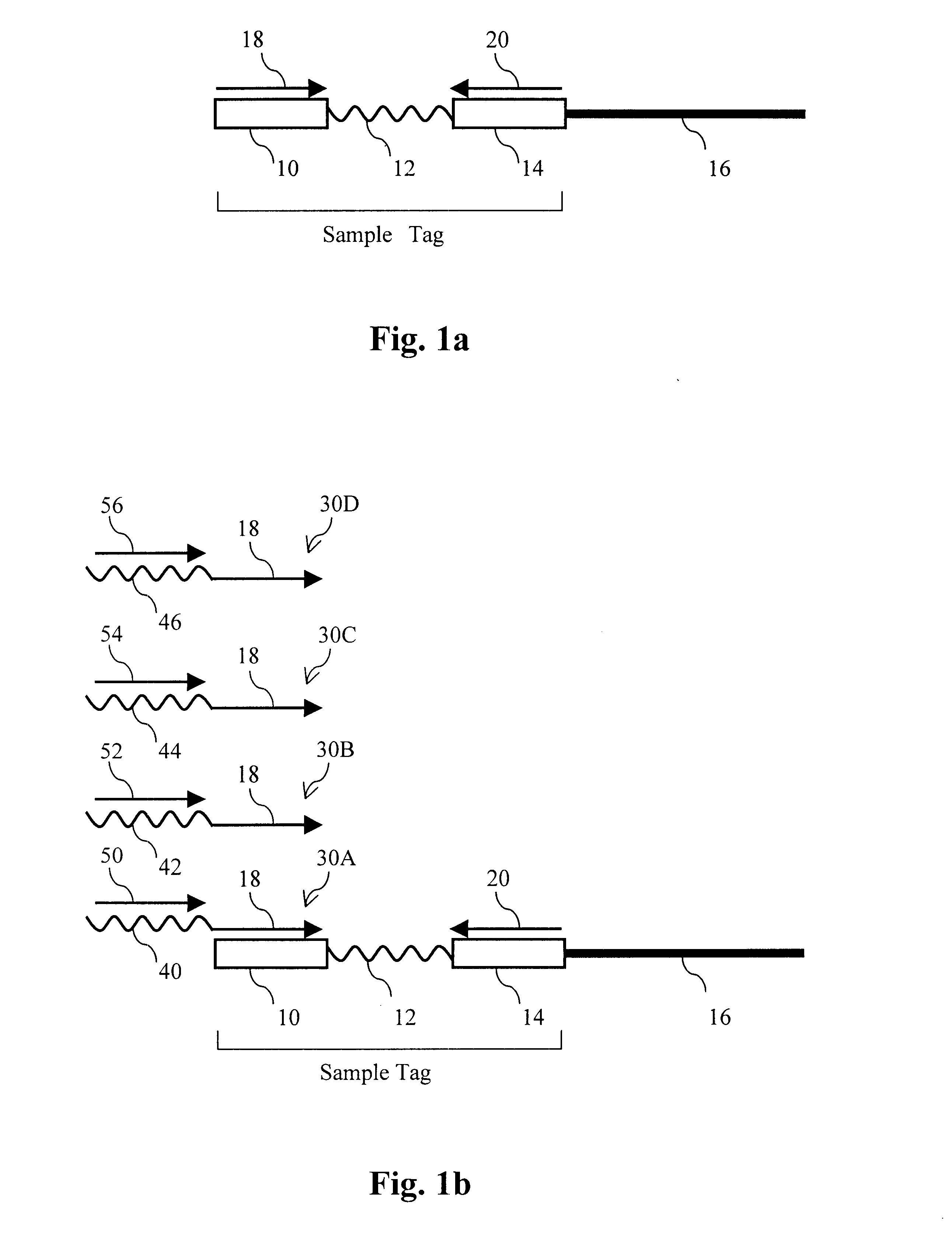
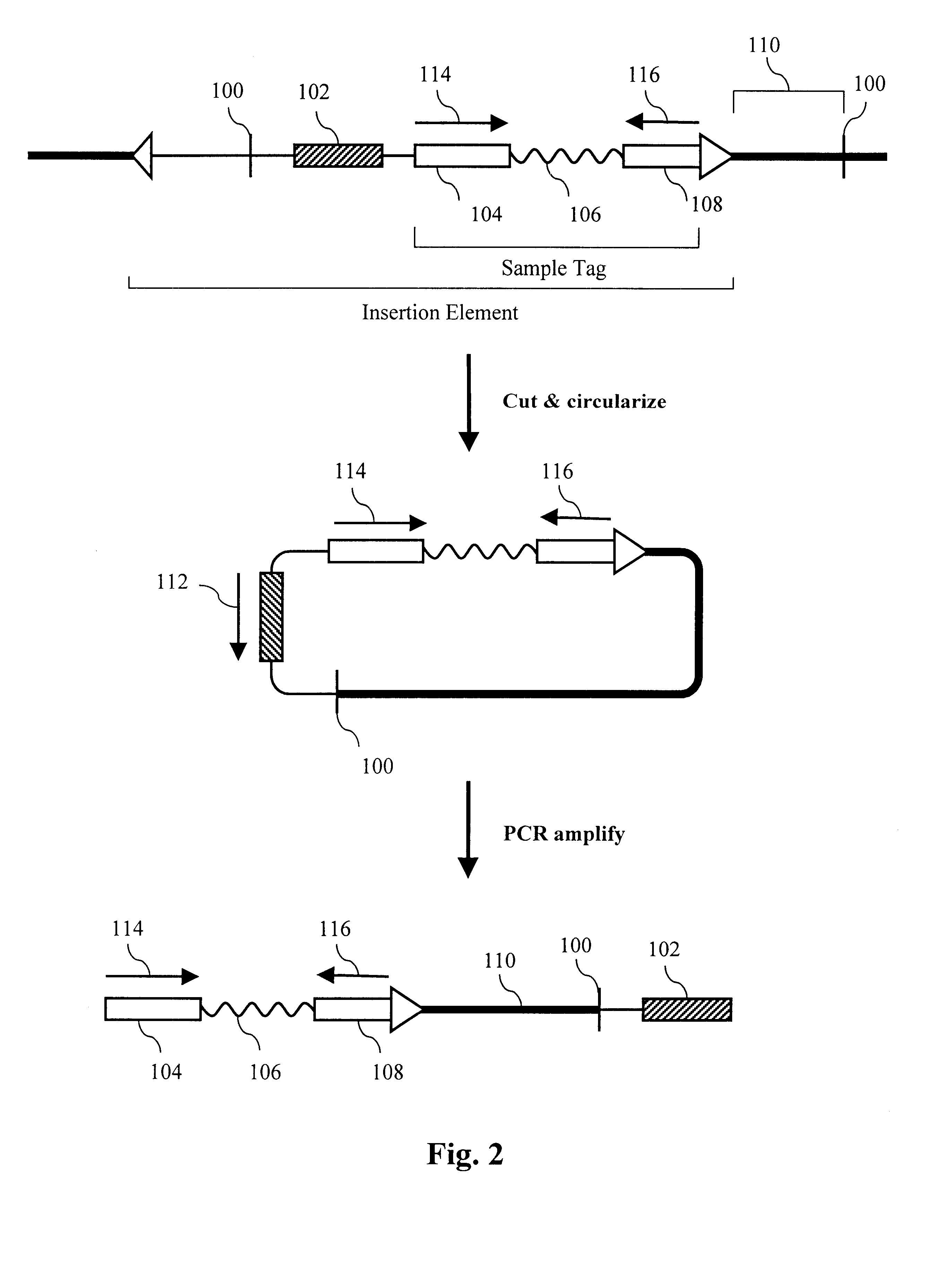
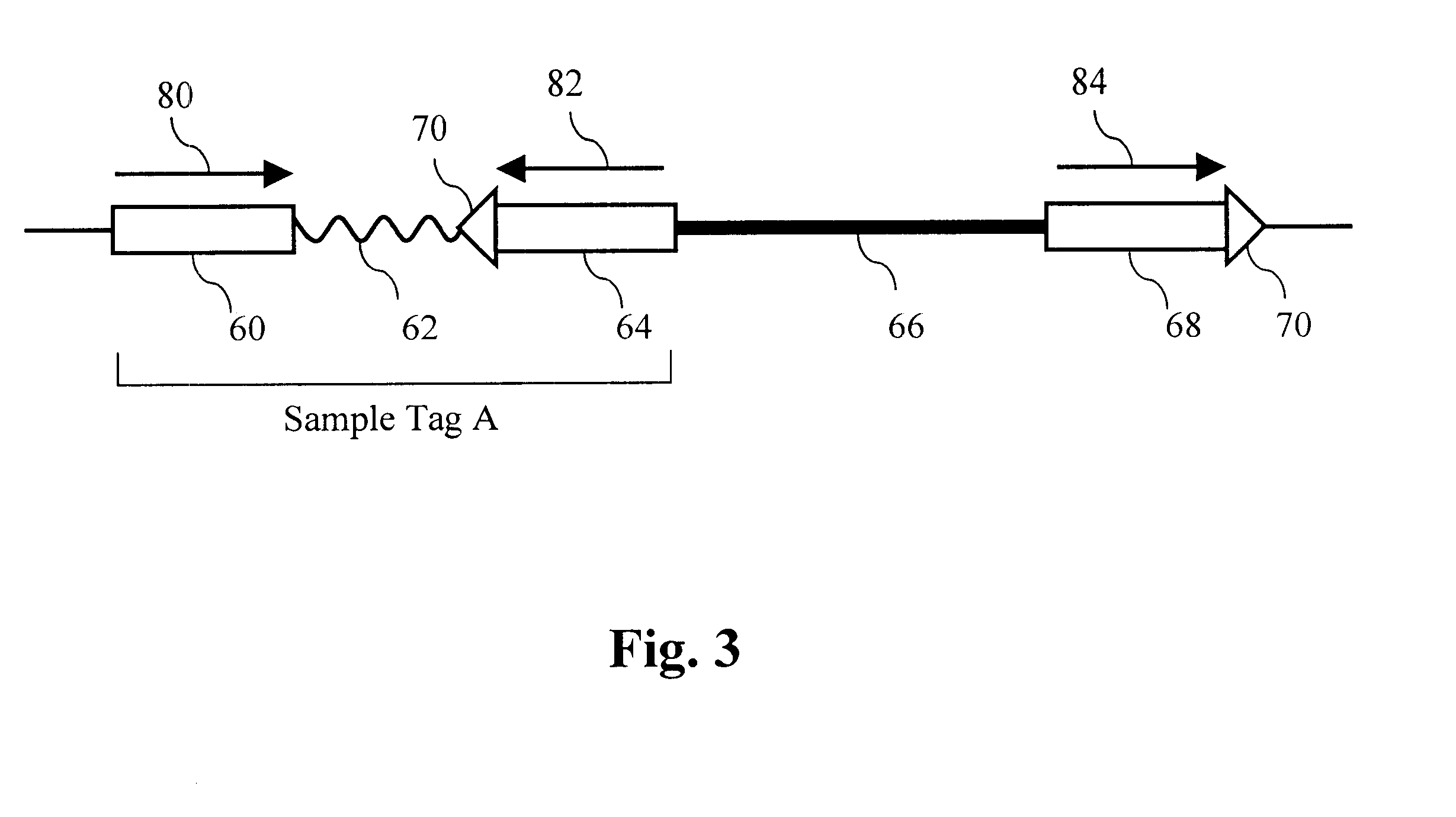
The present invention provides parallel methods for determining nucleotide sequences and physical maps of polynucleotides associated with sample tags. This information can be used to determine the chromosomal locations of sample-tagged polynucleotides. In one embodiment, the polynucleotides are derived from genomic DNA coupled to insertion elements. As a result, the invention also provides parallel methods for locating the integration sites of insertion elements in the genome.
Owner:STRATHMANN MICHAEL P
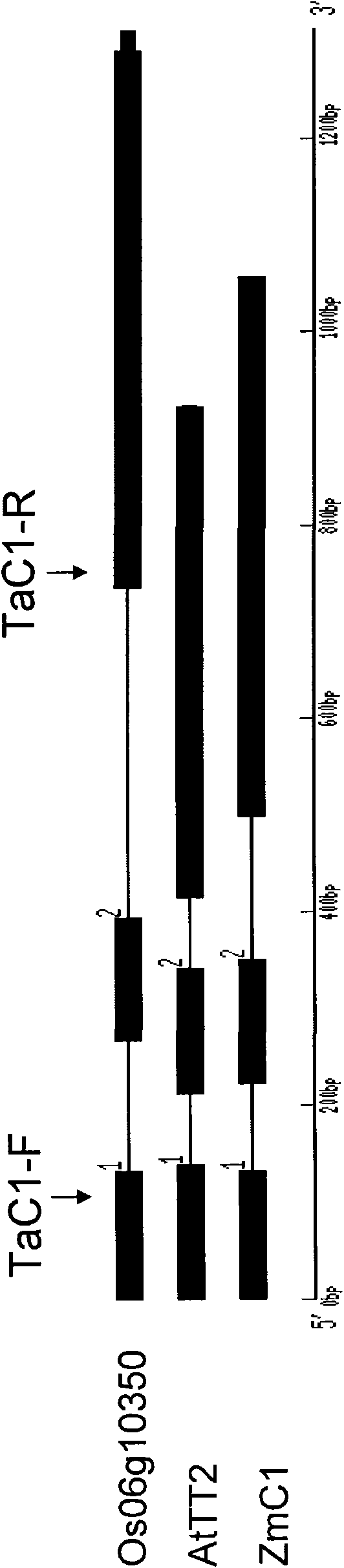
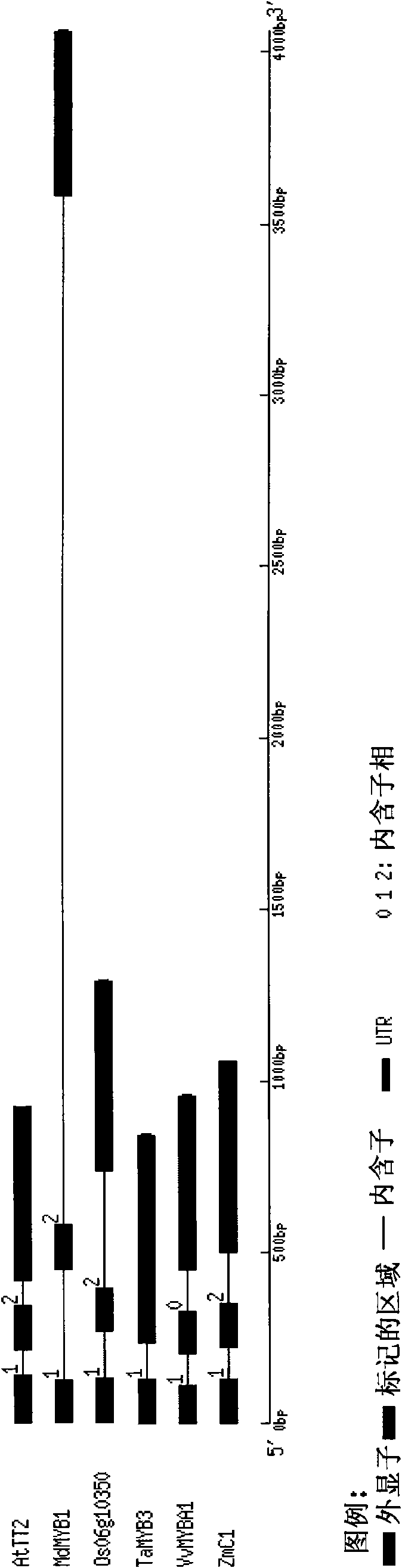
New wheat gene TaMYB3 for regulating synthetization and metabolization of anthocyanin
The invention provides a transcription factor TaMYB3 of a MYB class, which is separated from the plateau 115 of a purple kernel wheat variety and is used for regulating the synthetization and the metabolization of anthocyanin. TaMYB3 is positioned between 0.62 and 0.95 of a long arm physical map of wheat 4B chromosome. The subcellular fraction of a protein product is positioned on a cell nucleus. Shown by a derived amino acid sequence, the TaMYB3 codes a MYB class transcription factor regulating the synthetization and the metabolization of anthocyanin. The expression quantity of the TaMYB3 in the plateau 115 of a purple kernel wheat variety increases under the induction of light. The TaMYB3 can promote skin cells of the plateau 115 kernels after dark treatment to synthetize anthocyanin under the existence of transcription factors ZmR of corns bHLH during transient expression. Single TaMYB3 genes and single ZmR genes can not induct the synthetization of anthocyani, which shows that the TaMYB3 genes have the transcriptional activity of regulating the synthetization and the metabolization of anthocyanin. barly strip mosaic virus (BSMV) mediates the expression quantity decrease of the TaMYB3 in the plateau 115 kernels, and reduces the quantity of anthocyanin in kernels. This shows that the TaMYB3 participates in the biosynthesis of anthocyanin in the plateau 115 kernels.
Owner:INST OF GENETICS & DEVELOPMENTAL BIOLOGY CHINESE ACAD OF SCI +1
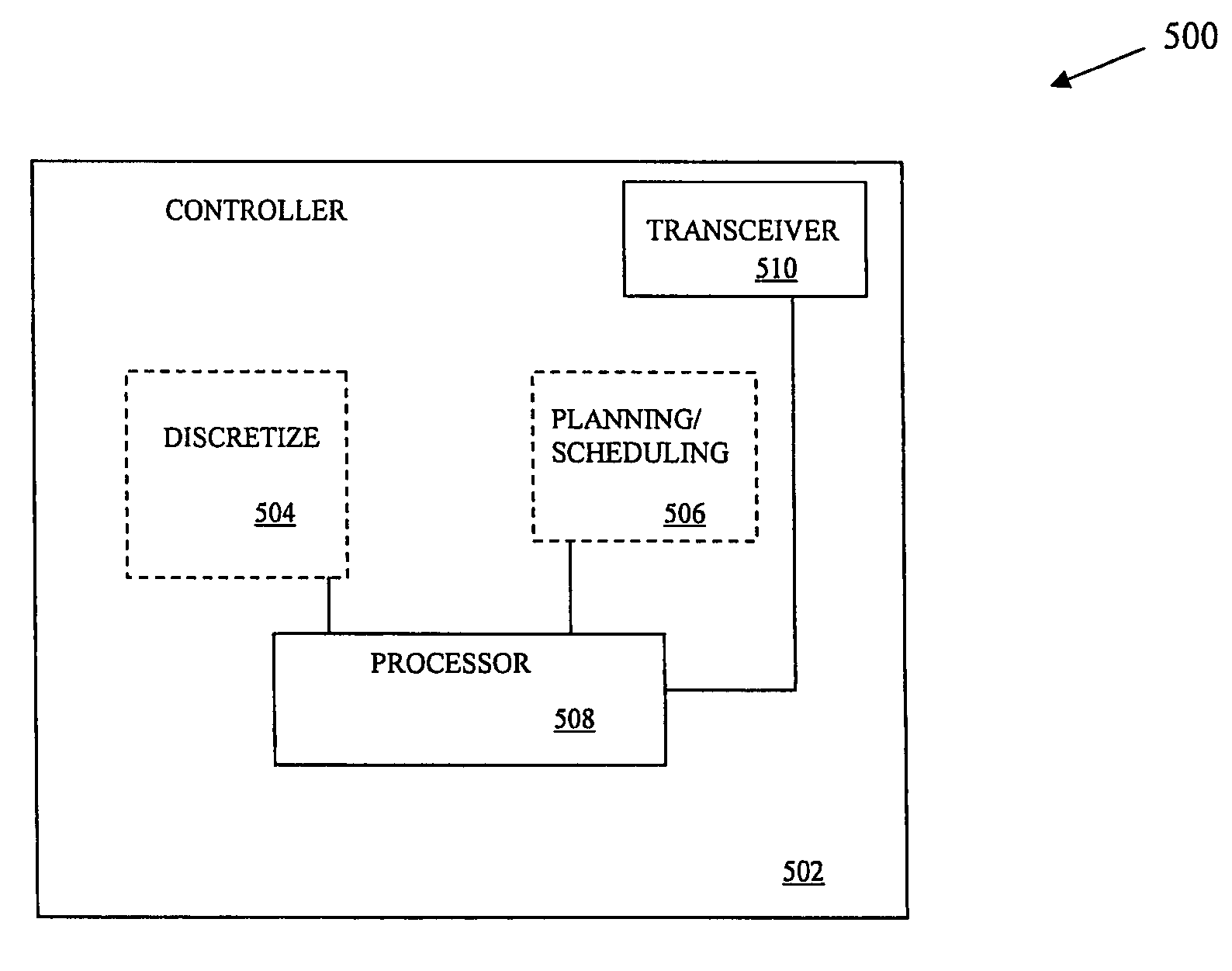
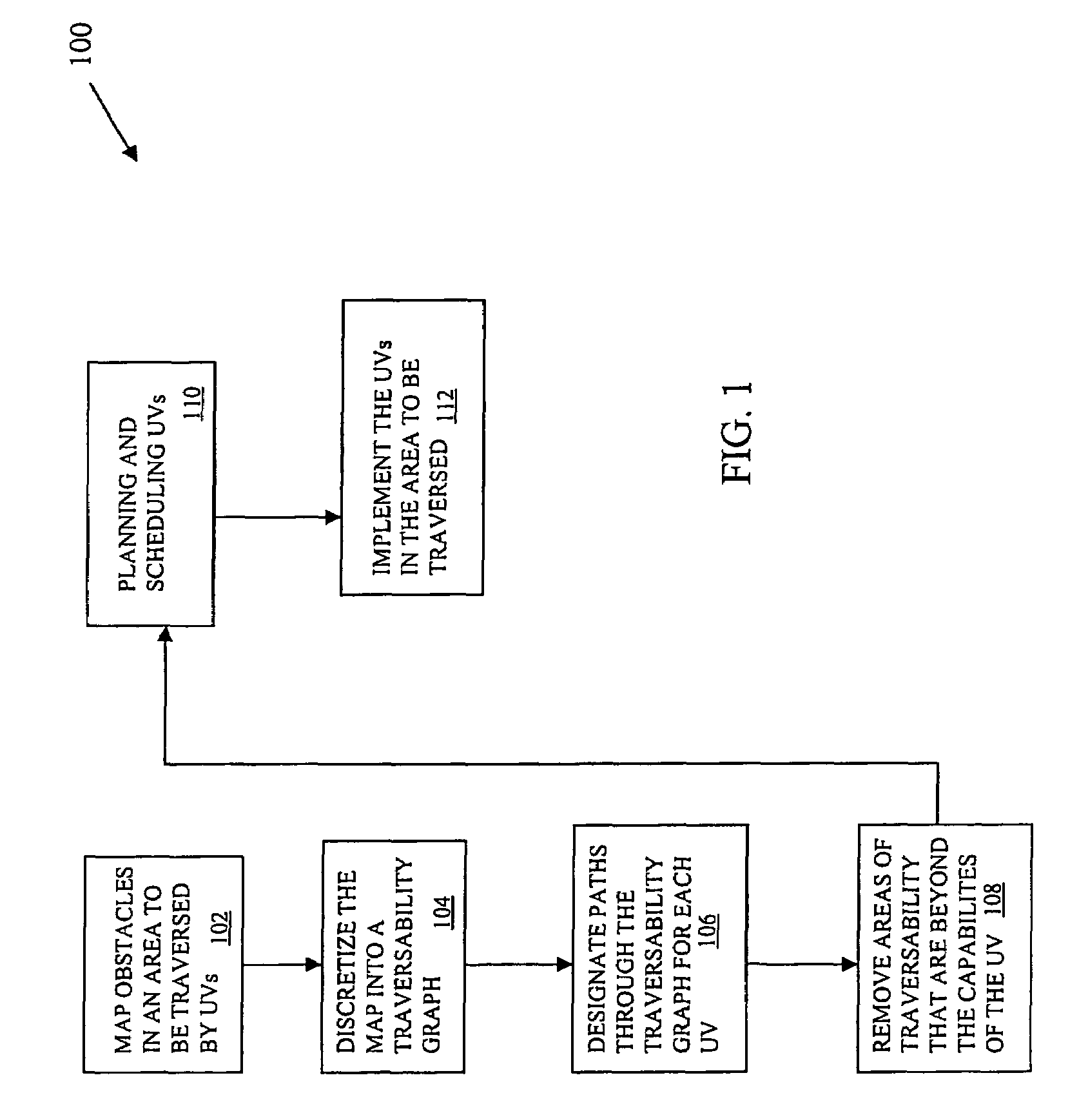
Real time planning and scheduling for a team of unmanned vehicles
Owner:HONEYWELL INT INC
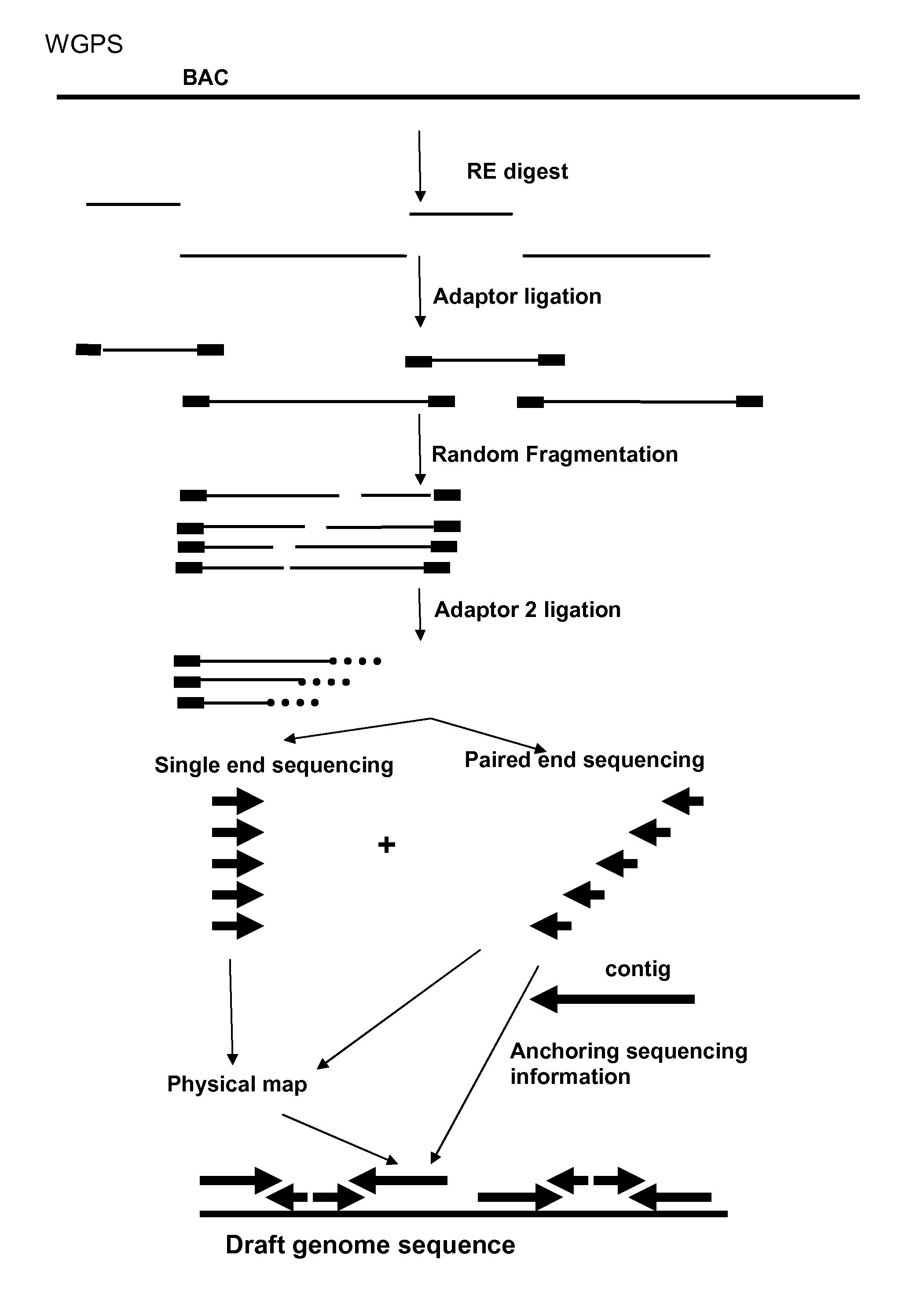
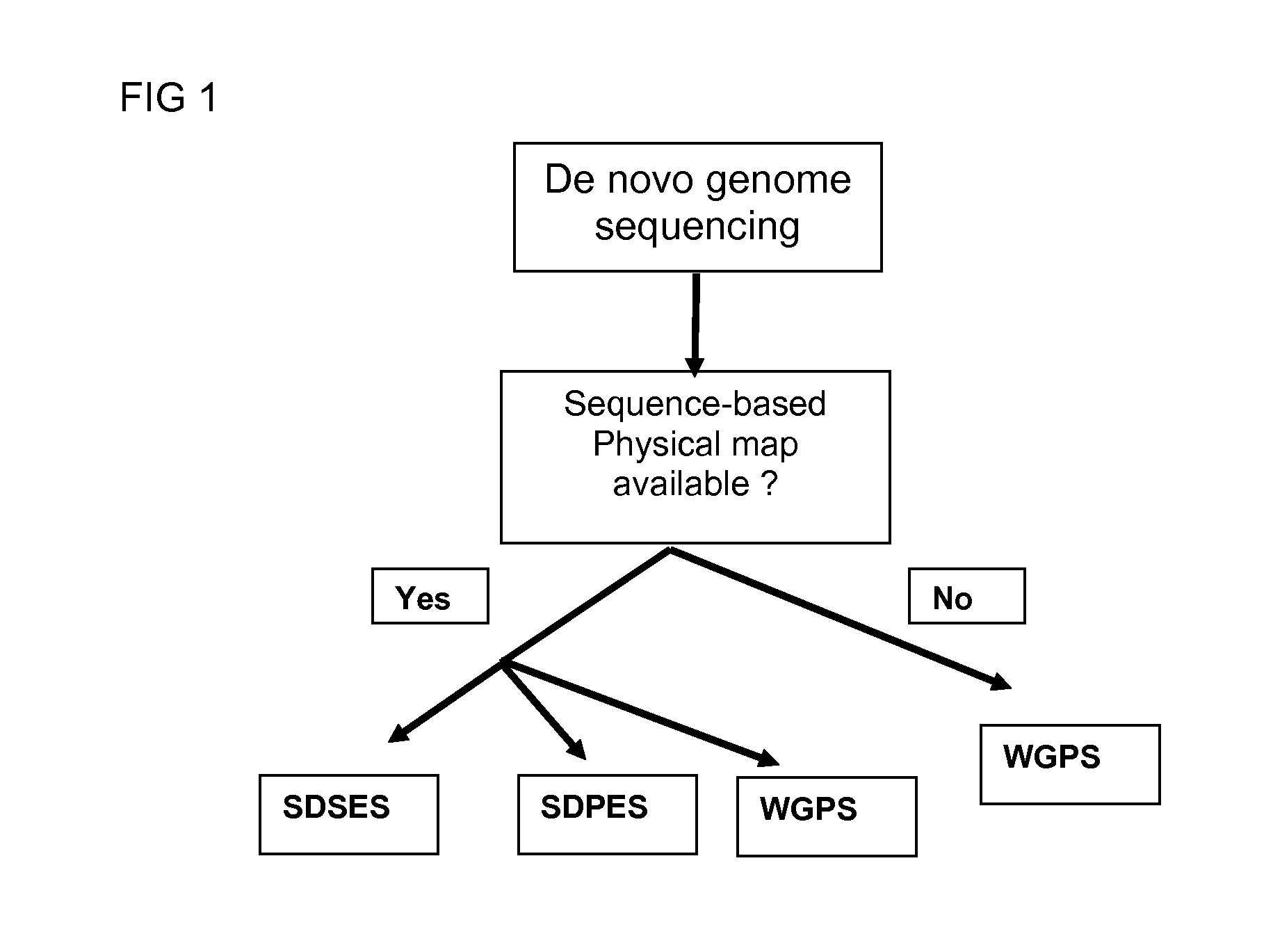
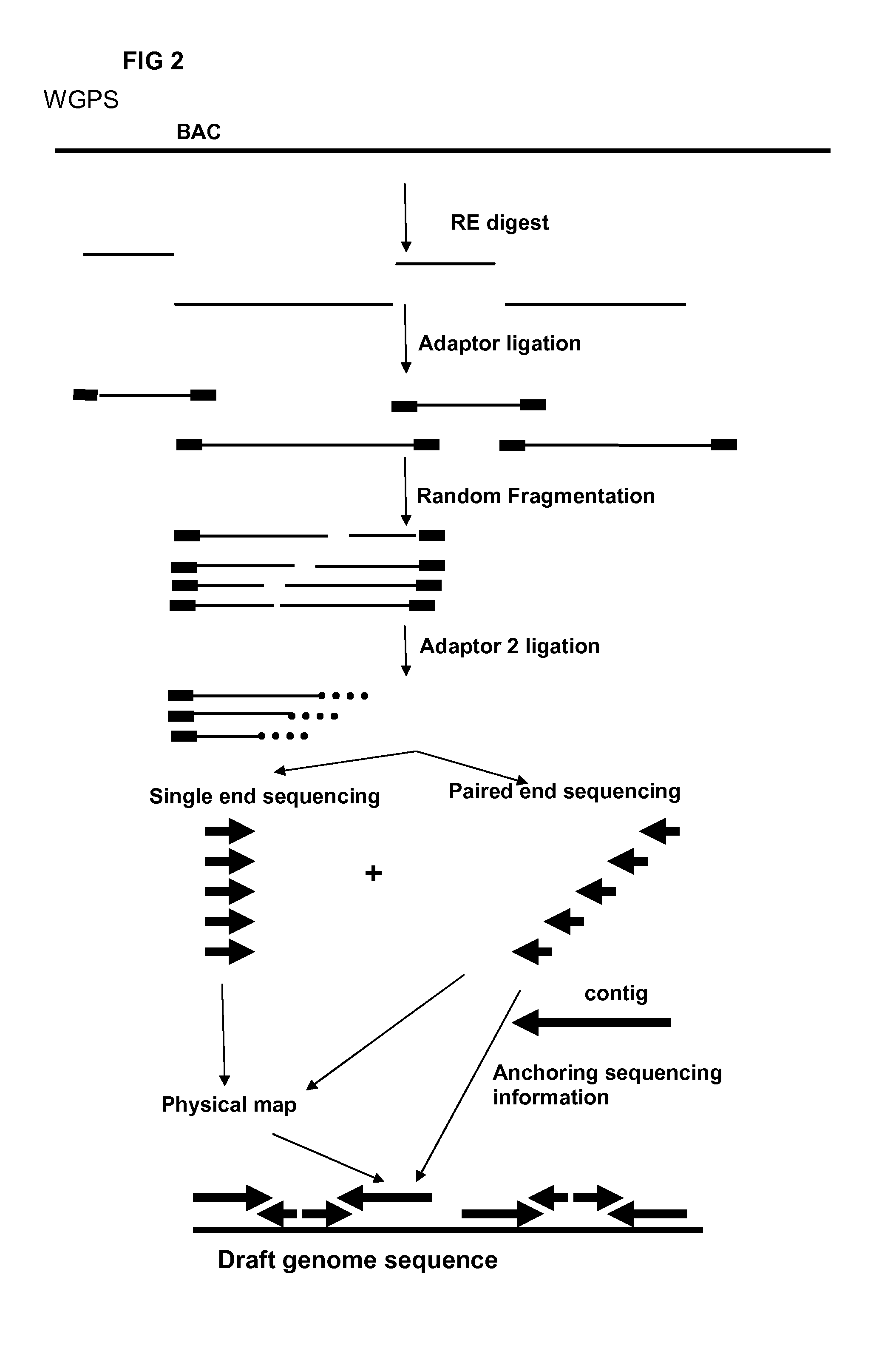
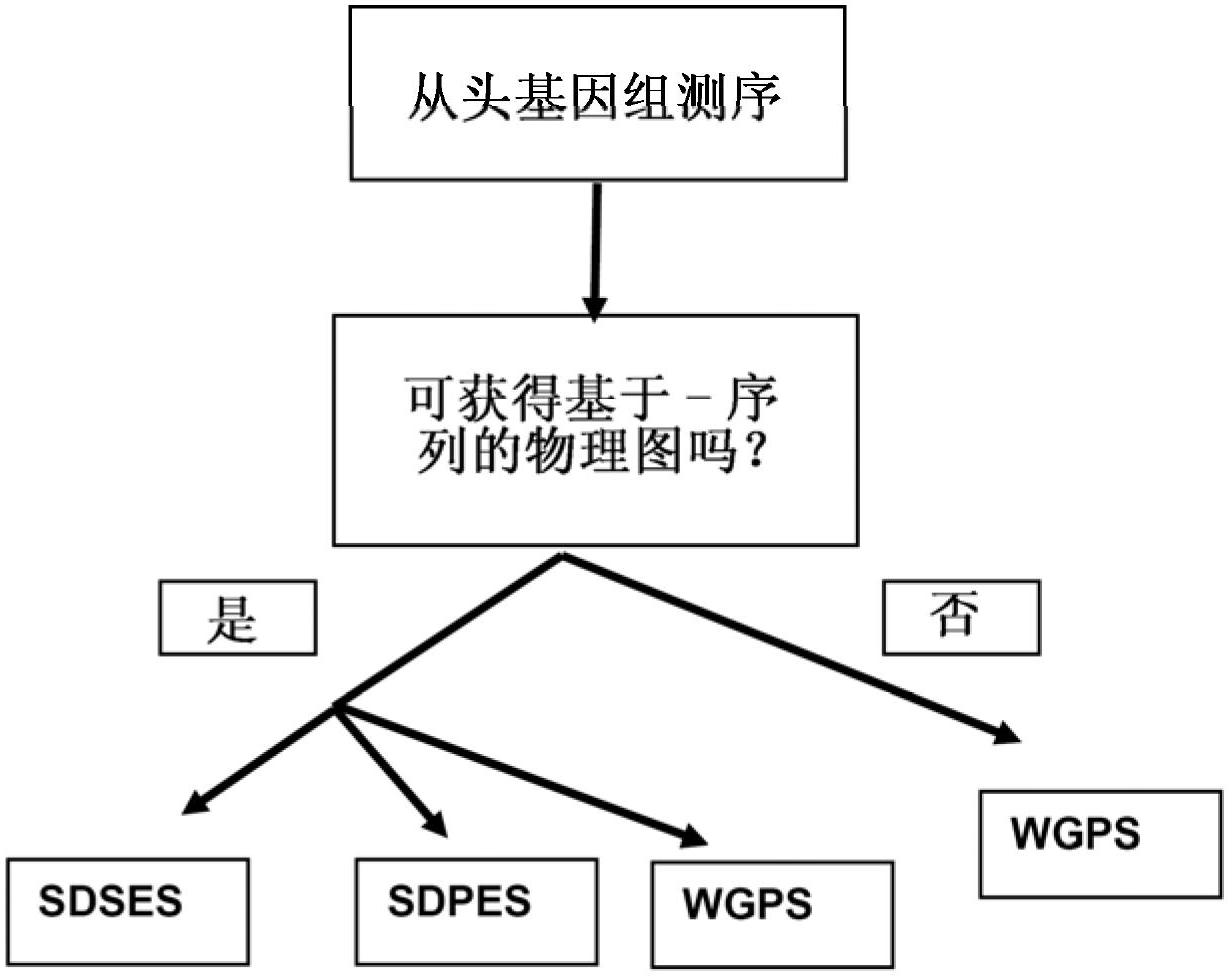
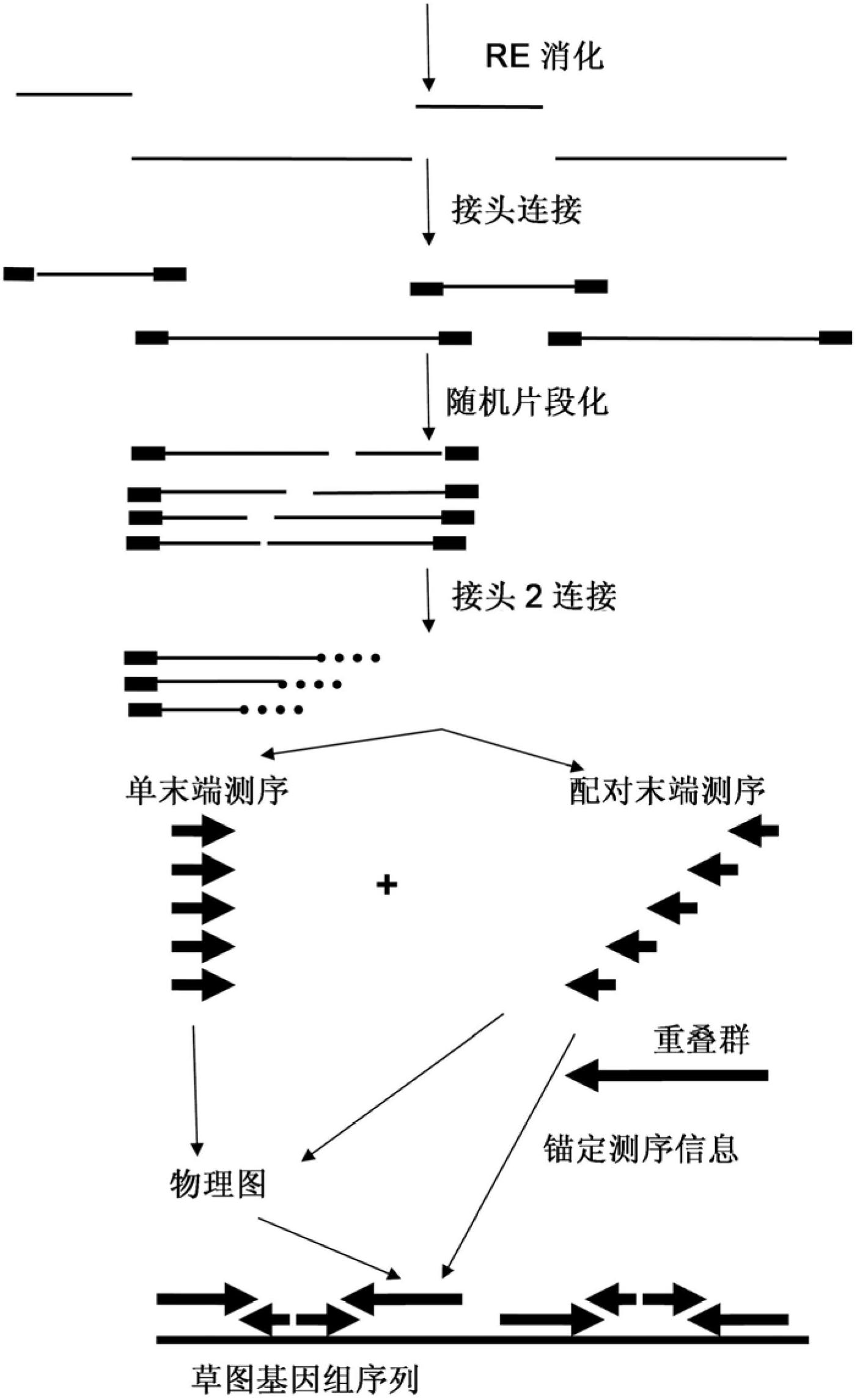
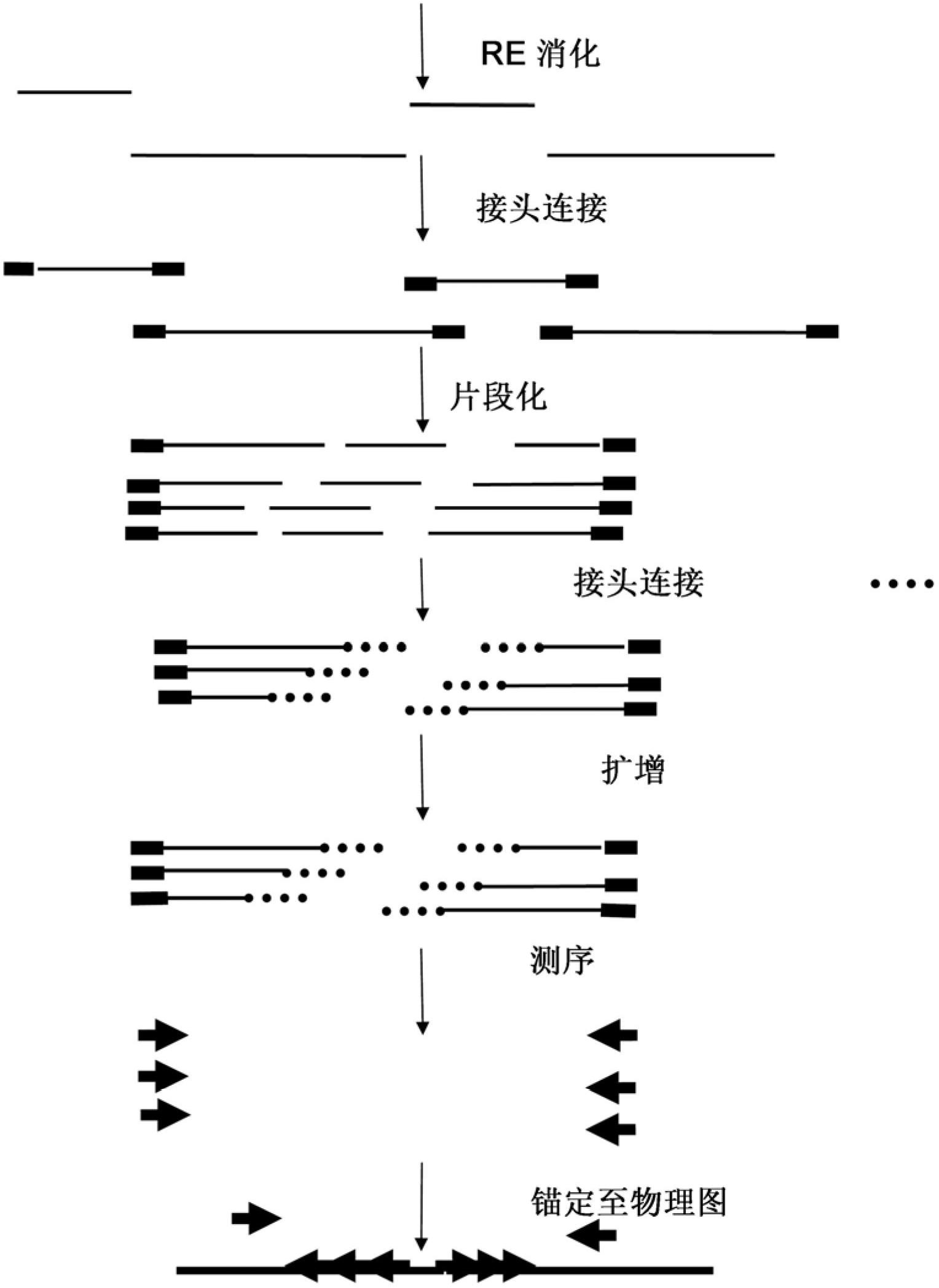
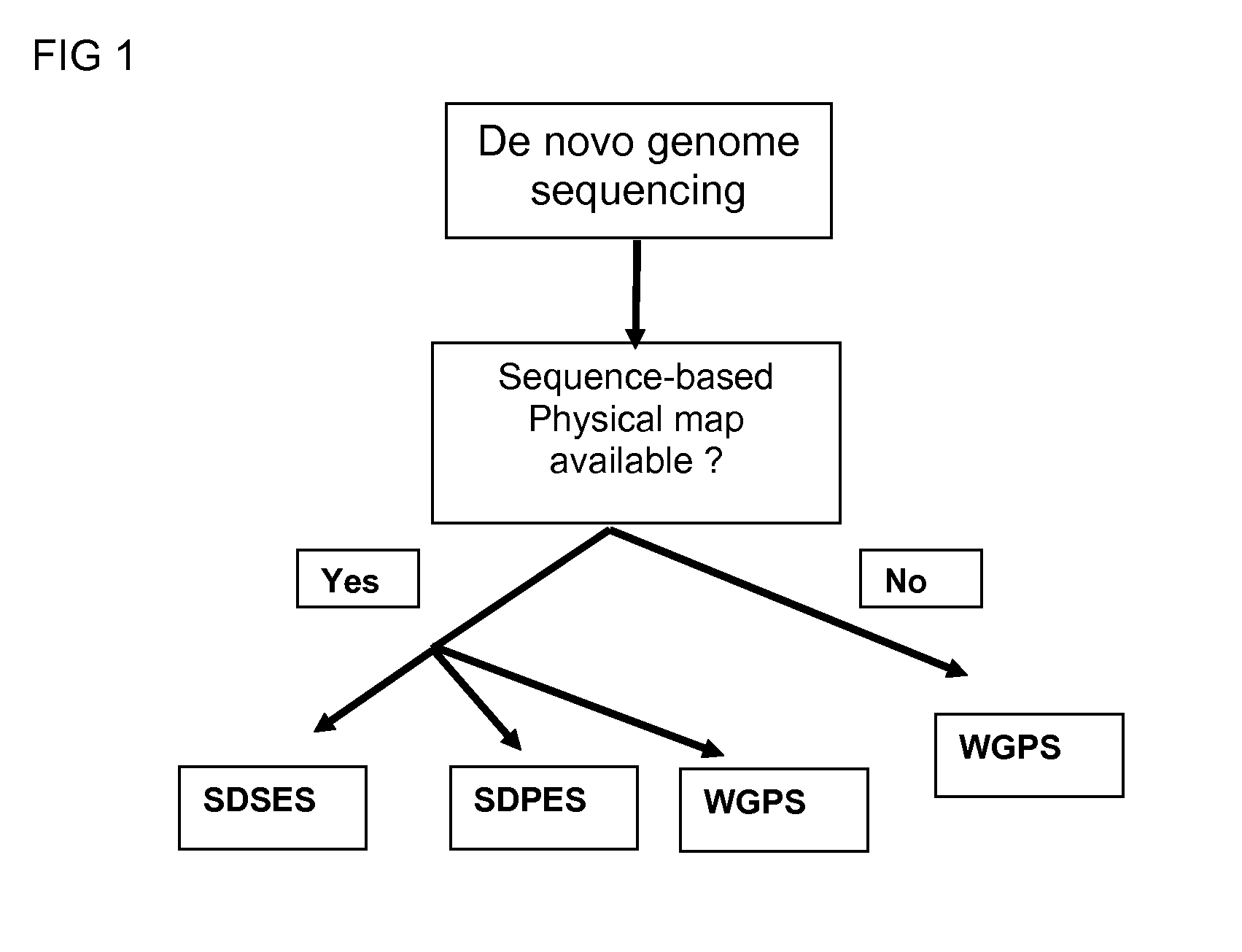
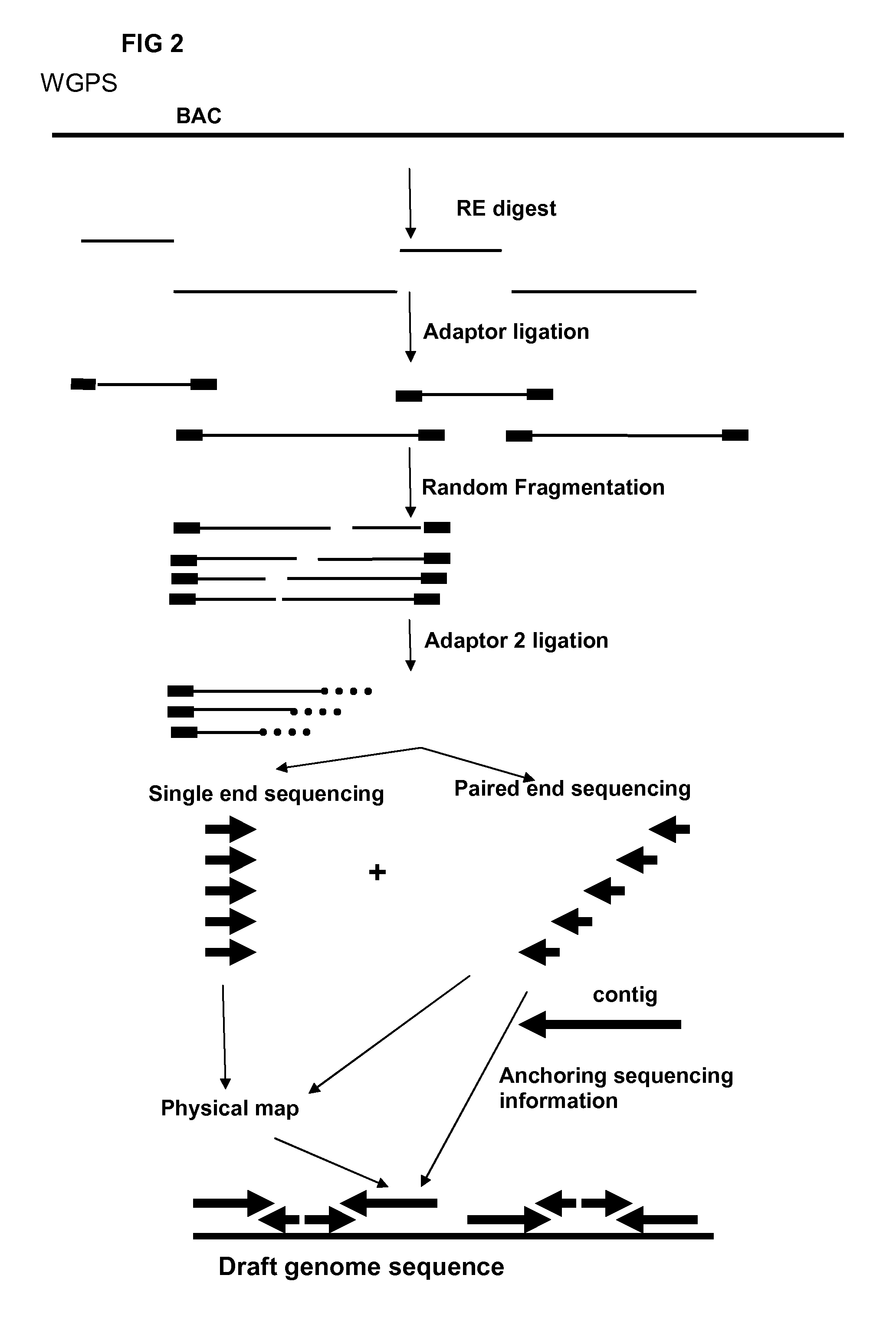
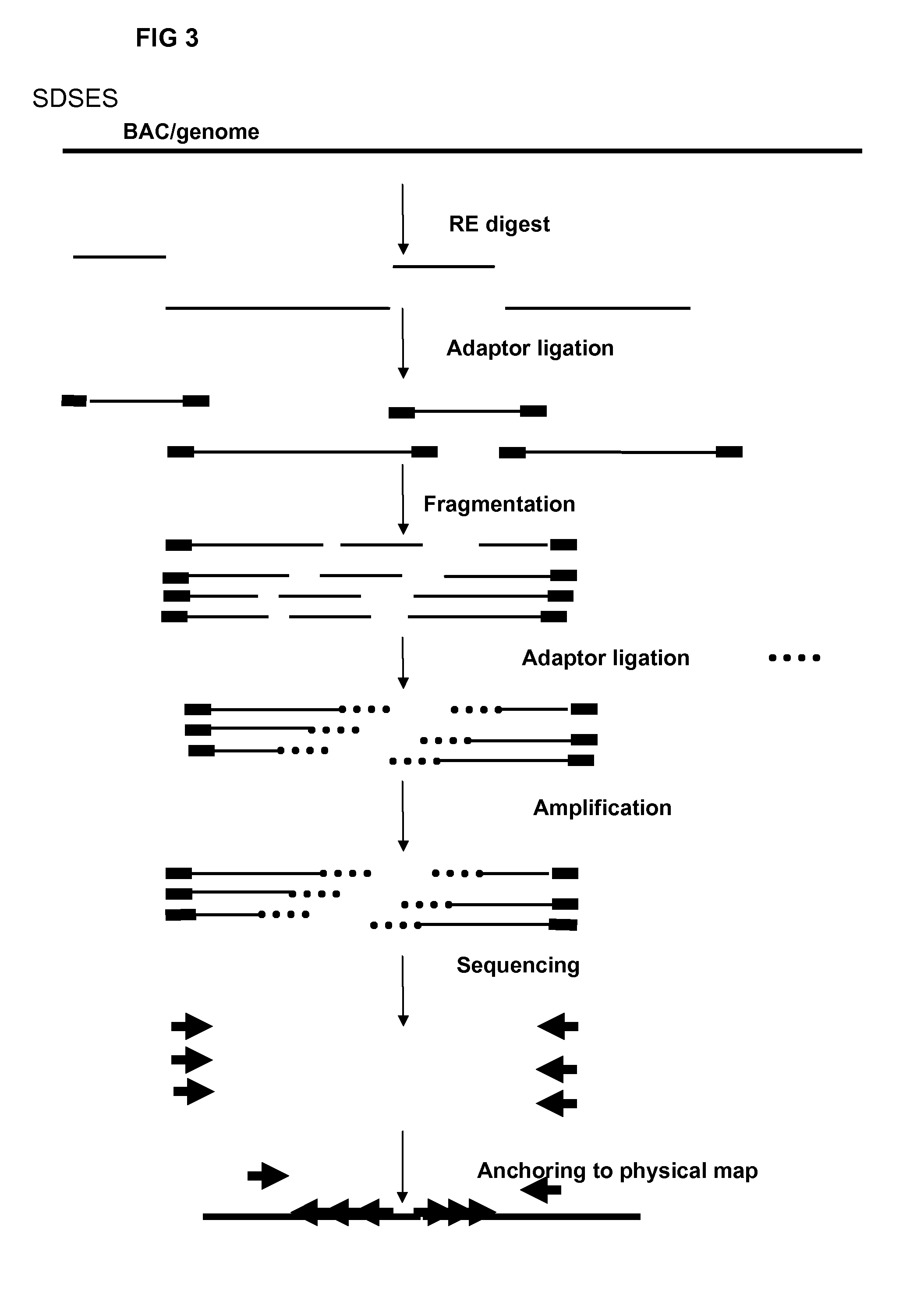
Restriction enzyme based whole genome sequencing
InactiveUS20120245037A1Maximize possibilityMicrobiological testing/measurementLibrary member identificationWhole genome sequencingPhysical Maps
Method for de novo whole genome sequencing based on a (sequence-based) physical map of a DNA sample clone bank based on end-sequencing tagged adapter-ligated restriction fragments, in combination with sequencing adapter-ligated restriction fragments of the DNA sample wherein the recognition sequence of the restriction enzyme used in the generation of the physical map is identical to at least part of the recognition sequence of the restriction enzyme used in the generation of the DNA sample.
Owner:KEYGENE NV
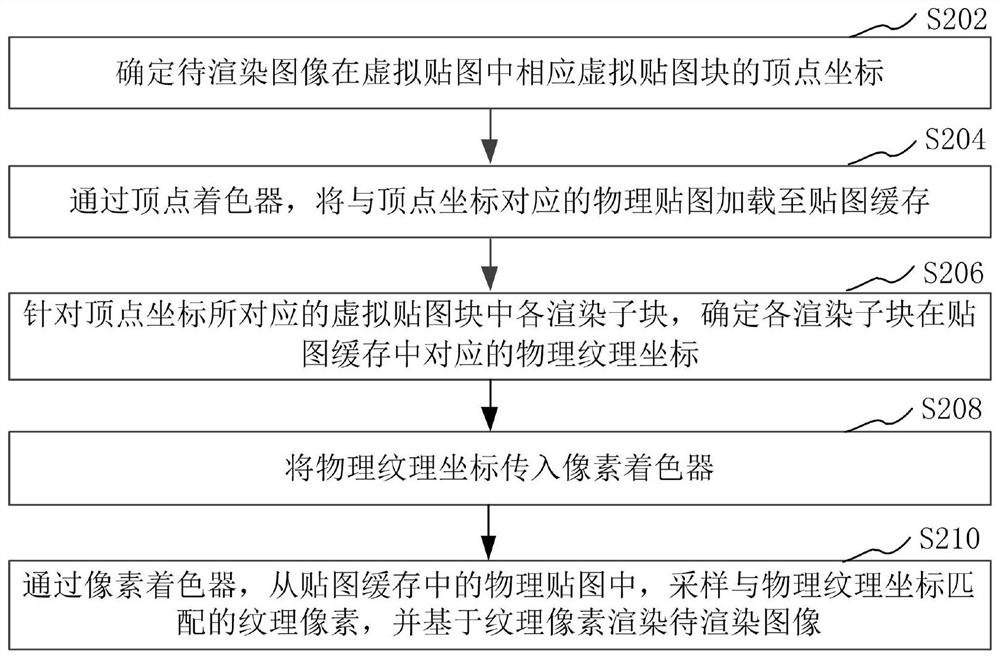
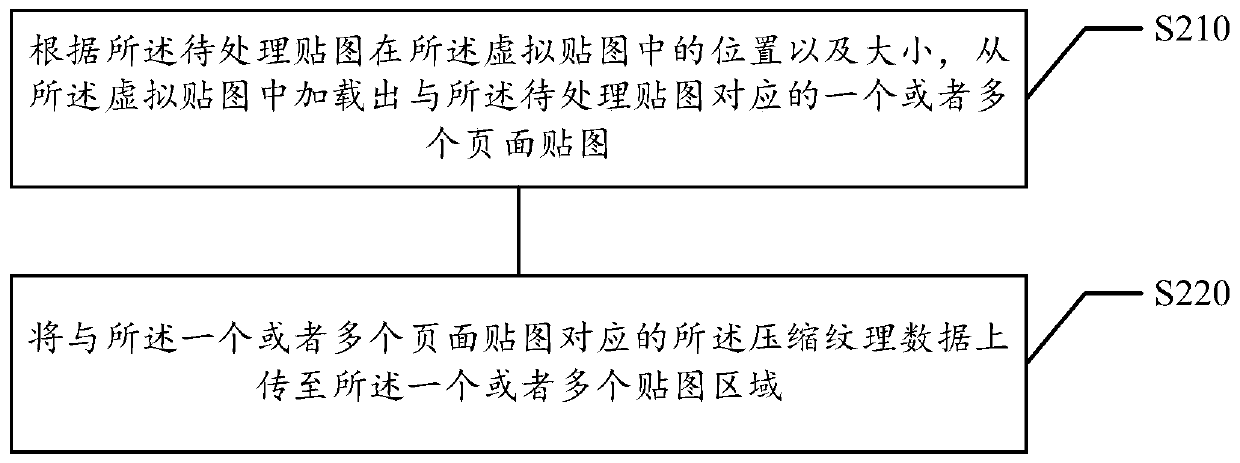
Image rendering method and device, computer equipment and storage medium
PendingCN112884875ASmall amount of calculationReduce resource consumptionImage enhancementImage analysisPattern recognitionComputer graphics (images)
The invention relates to an image rendering method and device, computer equipment and a storage medium. The method comprises the following steps: determining vertex coordinates of a corresponding virtual map block of an image to be rendered in a virtual map; loading a physical map corresponding to the vertex coordinate to a map cache through a vertex shader; for each rendering sub-block in a virtual map block corresponding to the vertex coordinate, determining a corresponding physical texture coordinate of each rendering sub-block in the map cache; transmitting the physical texture coordinates into a pixel shader; and through the pixel shader, sampling texture pixels matched with the physical texture coordinates from the physical map in the map cache, and rendering the to-be-rendered image based on the texture pixels. The resource consumption in the image rendering process can be effectively reduced, so that the image rendering efficiency is effectively improved.
Owner:TENCENT TECH (SHENZHEN) CO LTD
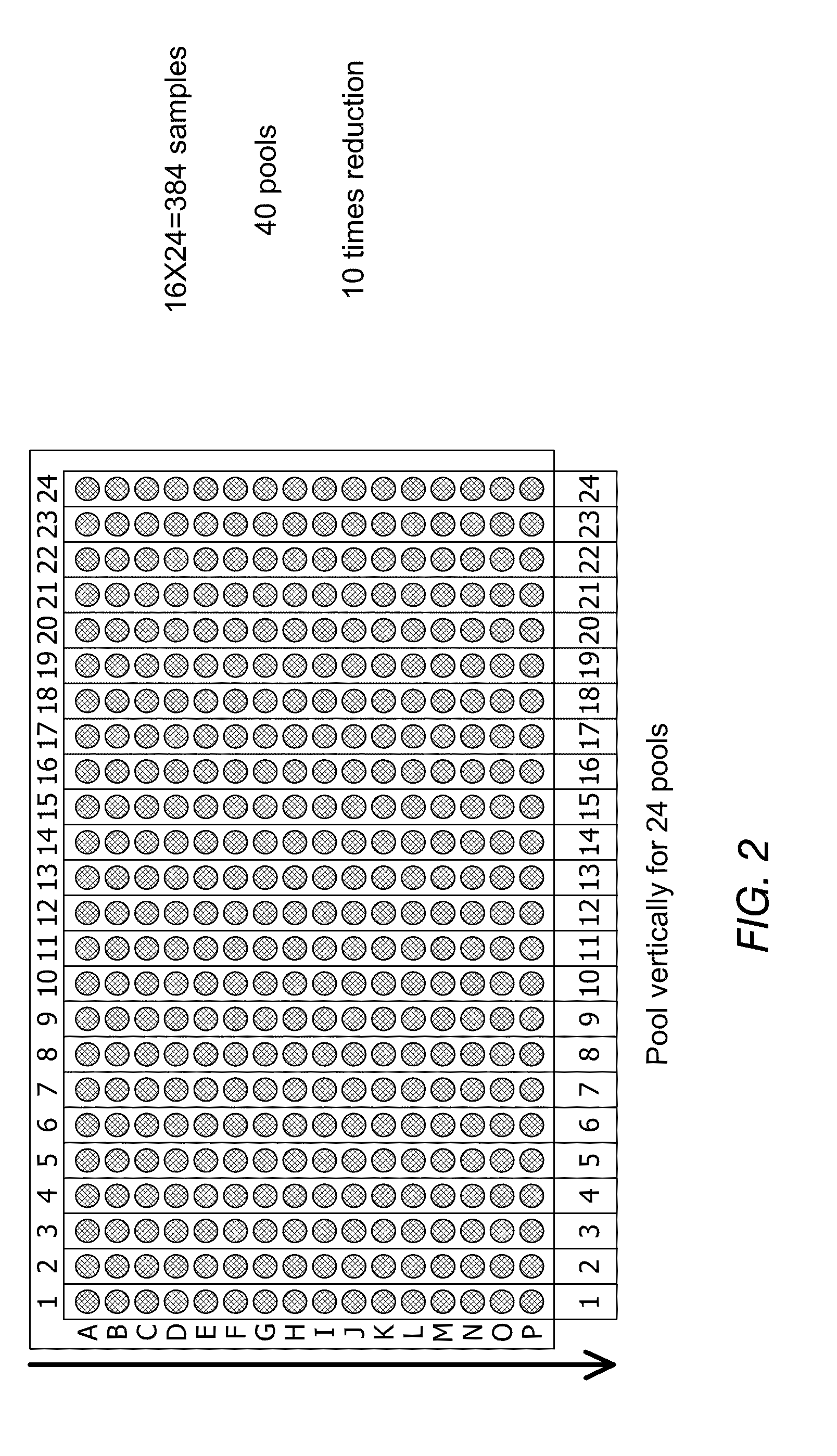
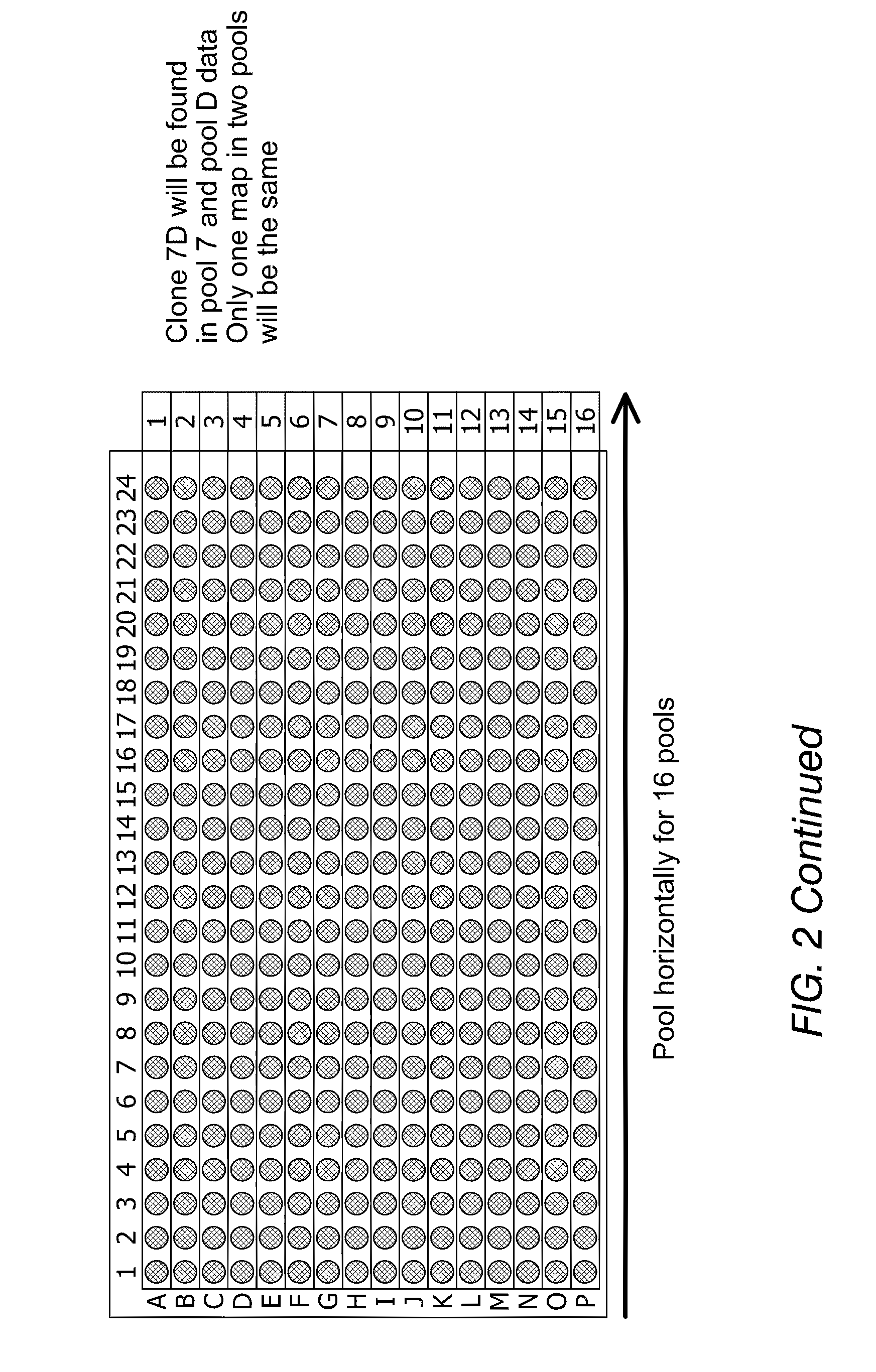
Physical map construction of whole genome and pooled clone mapping in nanochannel array
InactiveUS20130072386A1Improve throughputMicrobiological testing/measurementLibrary member identificationGenomic DNAPhysical Maps
Methods for generating physical maps for polynucleotides, such as genomic DNA, are disclosed herein. Also disclosed are methods for identifying the source of polynucleotides. The methods can, for example, be used in physical map construction of whole genome. In addition, methods and systems capable of performing high throughput characterization of macromolecules using nanofludic devices are enclosed.
Owner:BIONANO GENOMICS
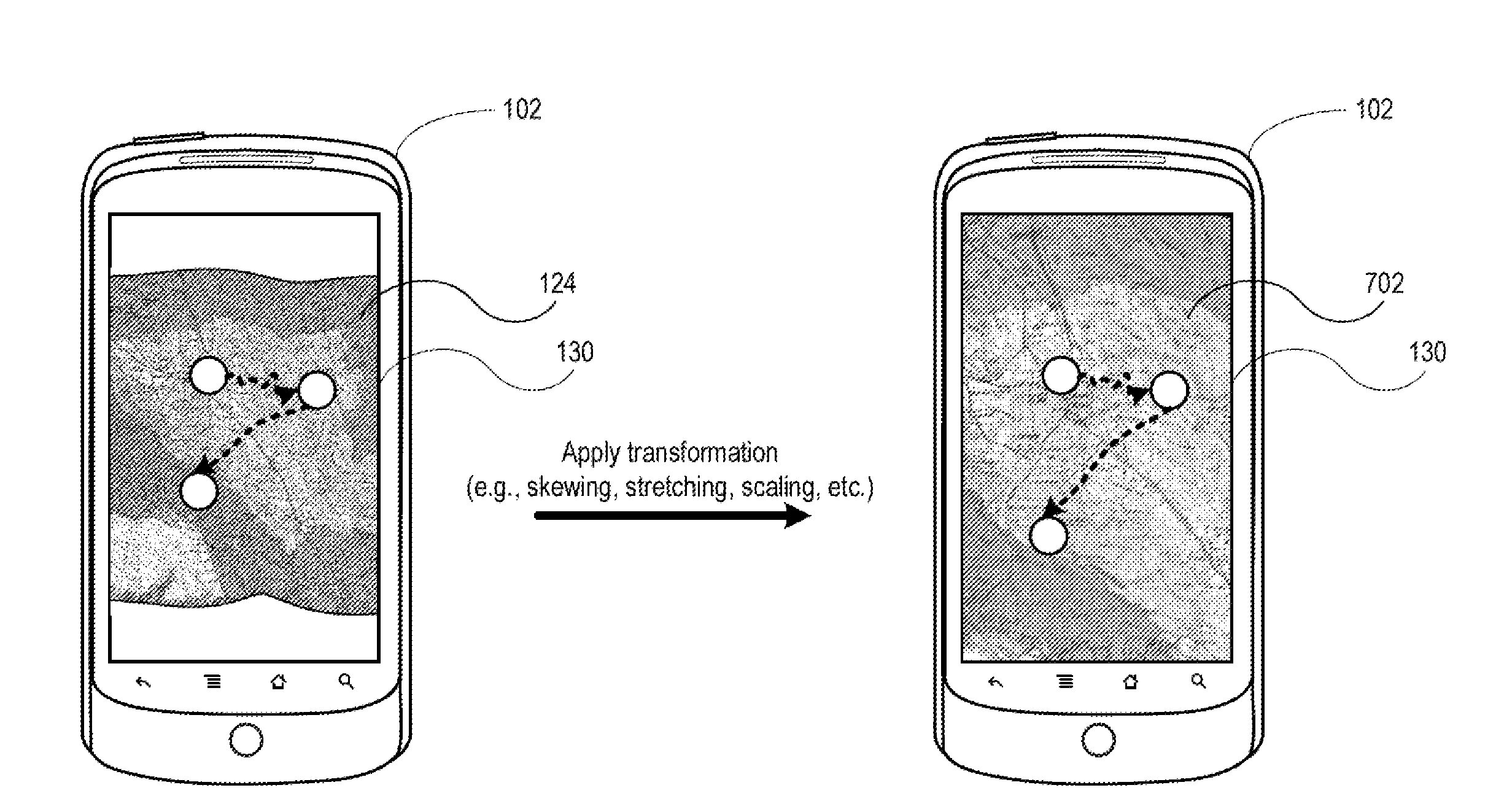
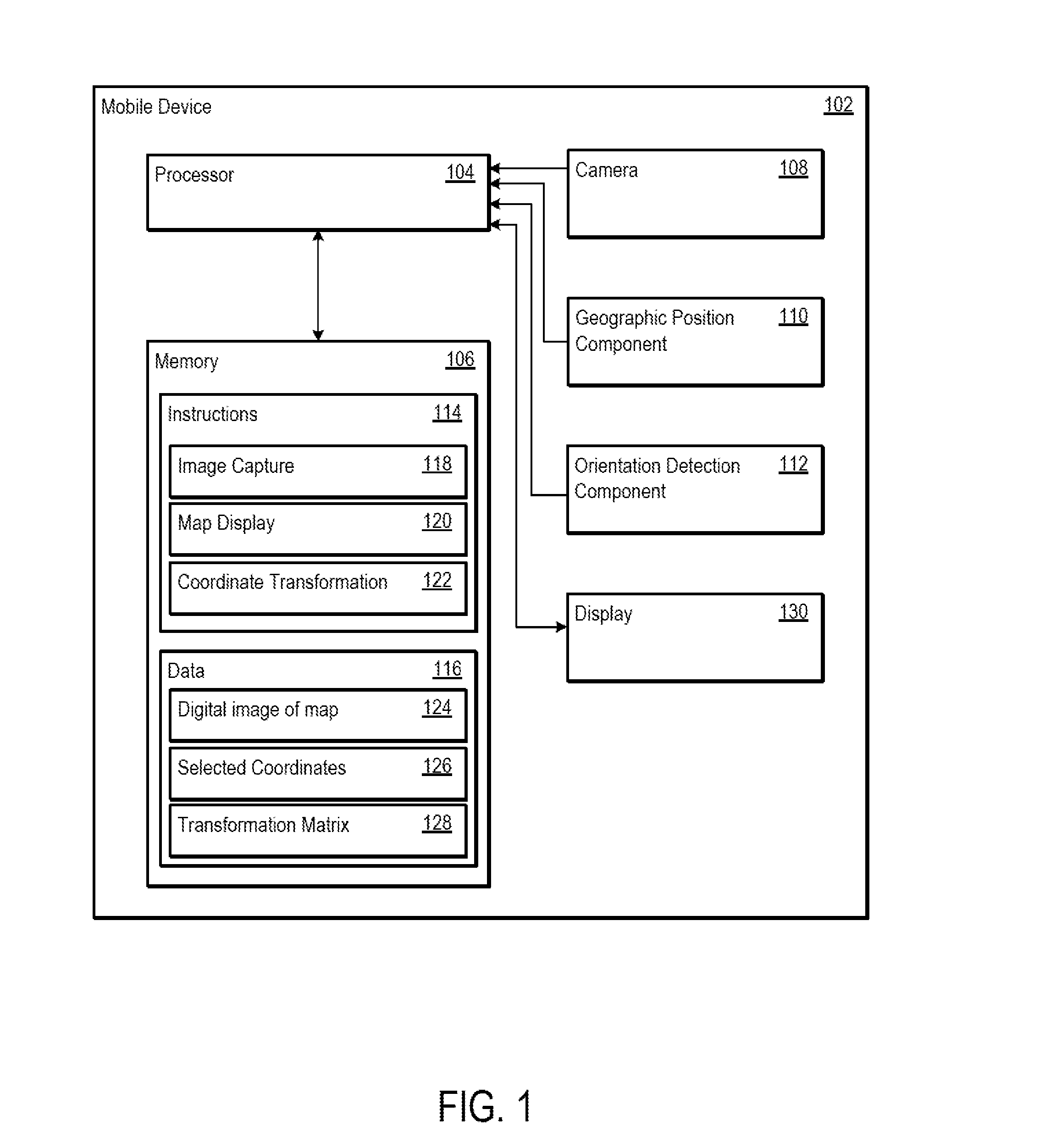
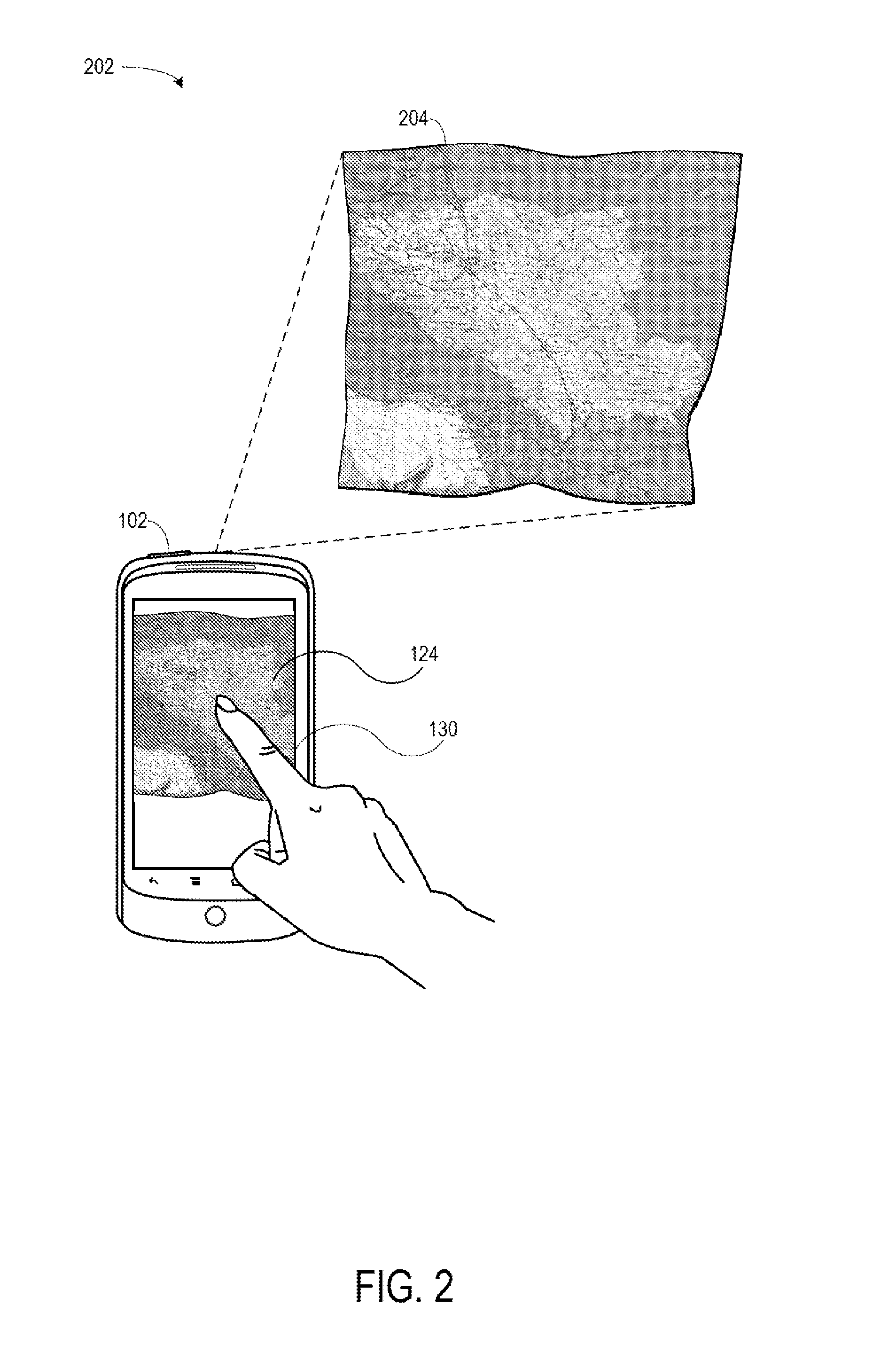
Using Geographic Coordinates On A Digital Image Of A Physical Map
InactiveUS20150154773A1Navigational calculation instrumentsCathode-ray tube indicatorsComputer graphics (images)Display device
Systems and methods are disclosed for using geographic coordinates on a physical map. The system may include a mobile device having a processor and a display, where the processor receives a first input that instructs a camera to capture a digital image of a physical map. The mobile device may then display the digital image of the physical map. The mobile device may further receive a second input from a user interface, where the second input identifies a first location on the digital image of the physical map. The first location may be associated with an image pixel coordinate, and the mobile device may obtain a geographic coordinate based on this image pixel coordinate. The mobile device may then determine a transformation matrix based on the image pixel and geographic coordinates. Using the determined transformation matrix, the mobile device may determine further image pixel coordinates based on received geographic coordinates.
Owner:GOOGLE LLC
Restriction enzyme based whole genome sequencing
A method for de novo whole genome sequencing based on a (sequence-based) physical map of a DNA sample clone bank based on end-sequencing tagged adapter-ligated restriction fragments, in combination with sequencing adapter-ligated restriction fragments of the DNA sample wherein the recognition sequence of the restriction enzyme used in the generation of the physical map is identical to at least part of the recognition sequence of the restriction enzyme used in the generation of the DNA sample.
Owner:KEYGENE NV
Novel genome sequencing strategies
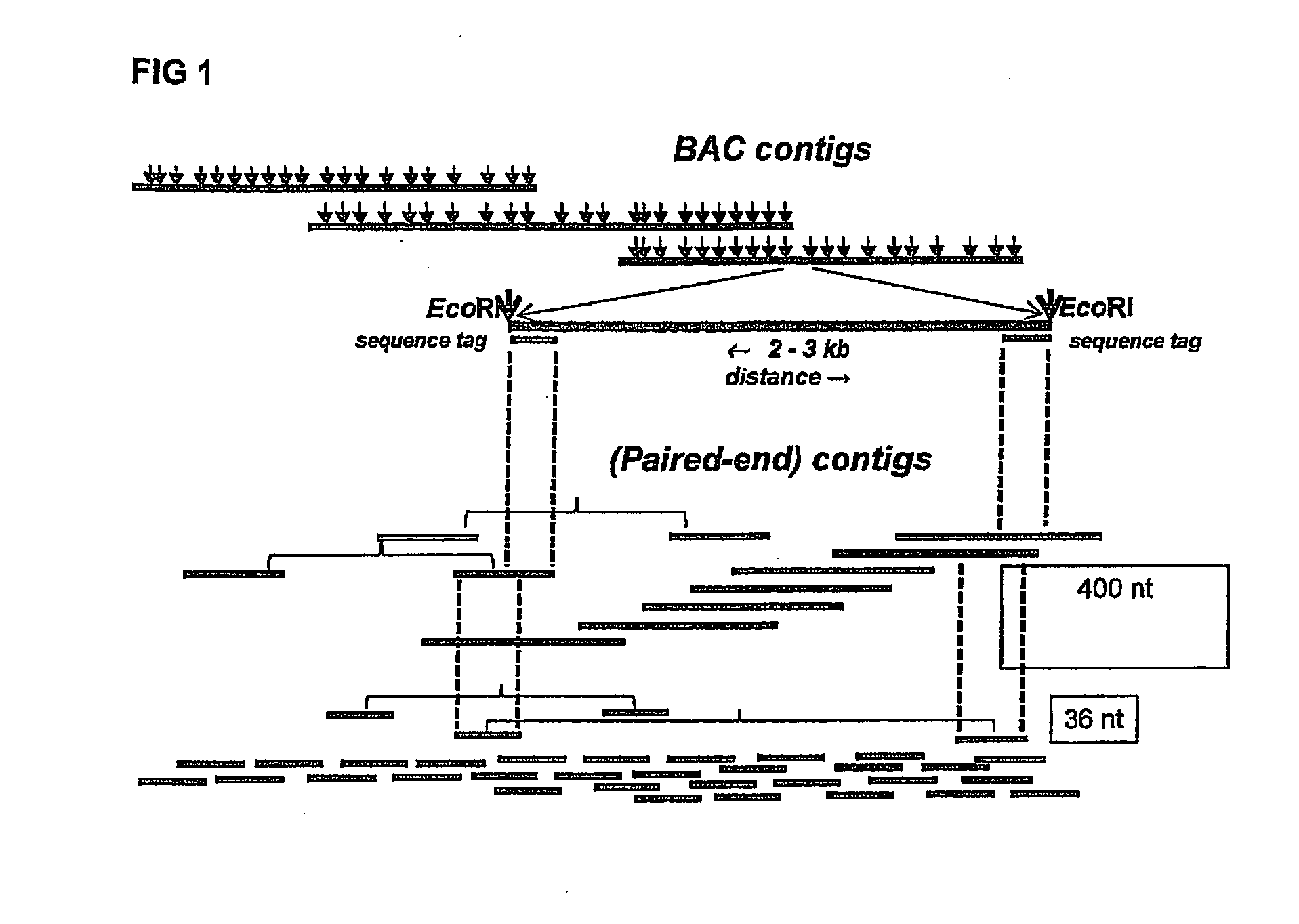
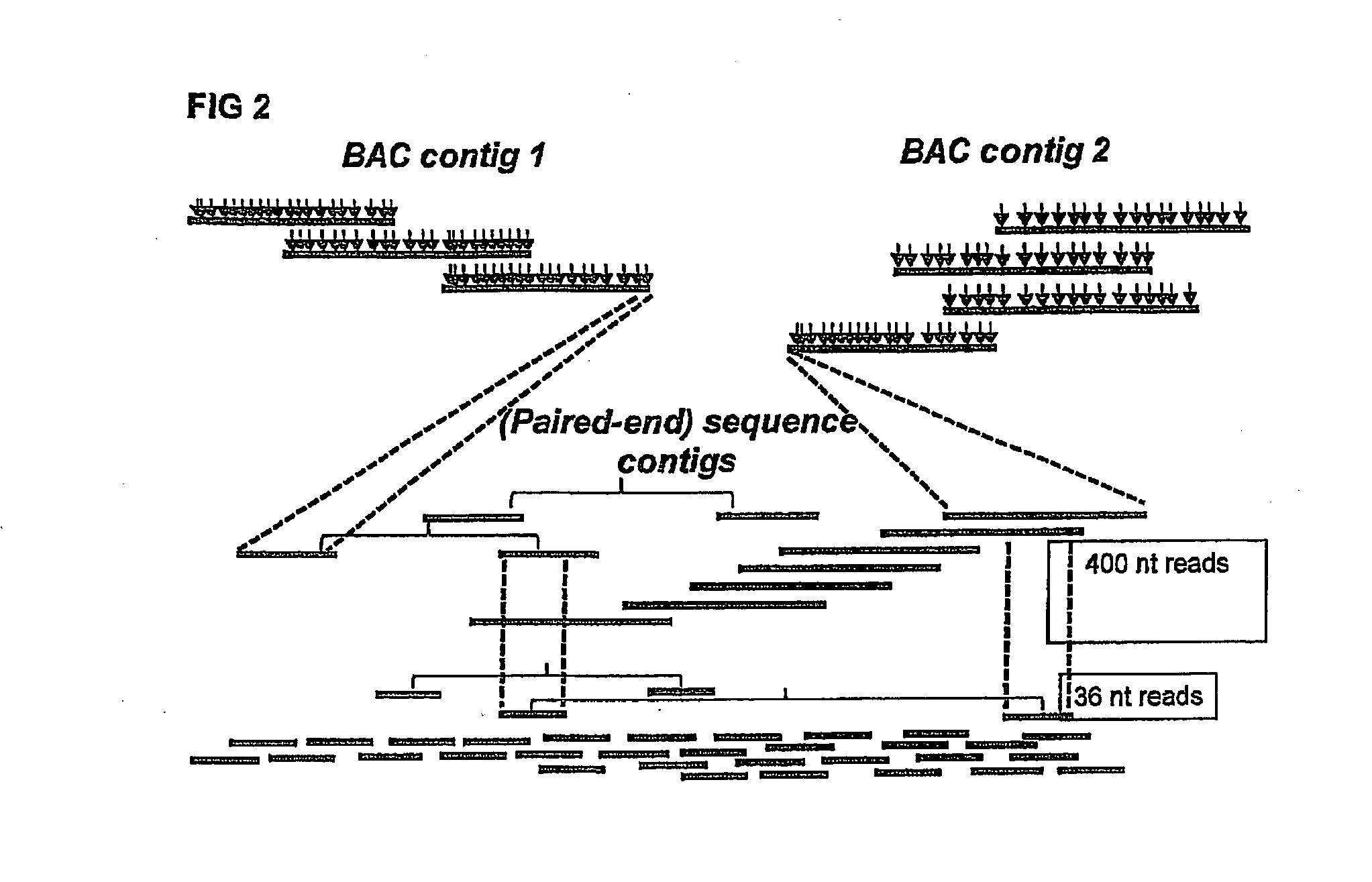
InactiveUS20130196859A1Increased length and densityQuality improvementMicrobiological testing/measurementLibrary member identificationBac cloneGenomic sequencing
The invention relates to a method for the determination of a genome sequence comprising the steps of providing a physical map of a sample genome by sequencing fragment ends of pooled BAC clones; providing a set of sequence reads from a sample genome generating a contig of the physical map and the sequence reads.
Owner:KEYGENE NV
Sensor data collection
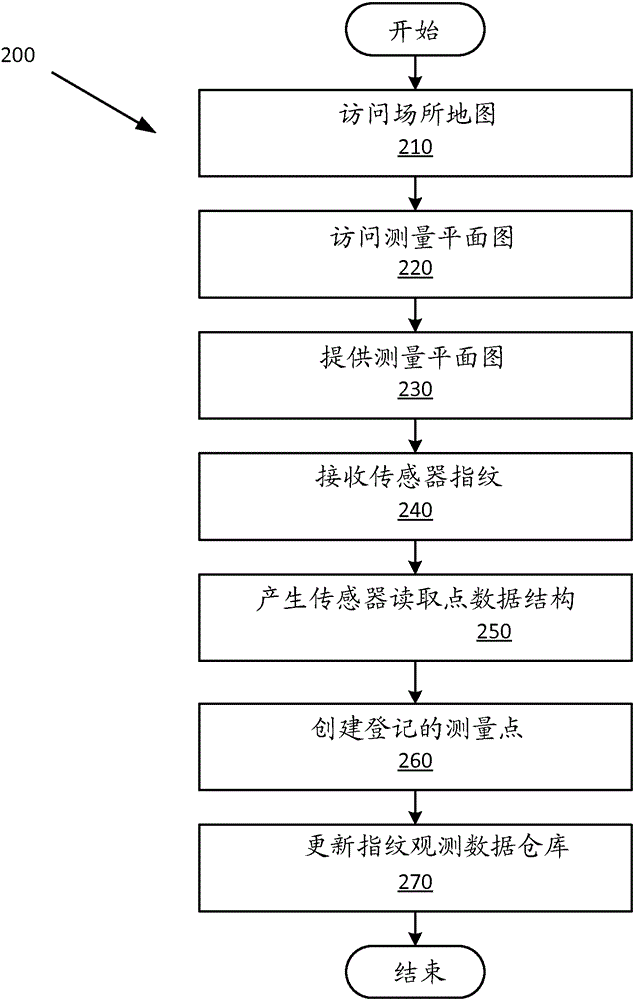
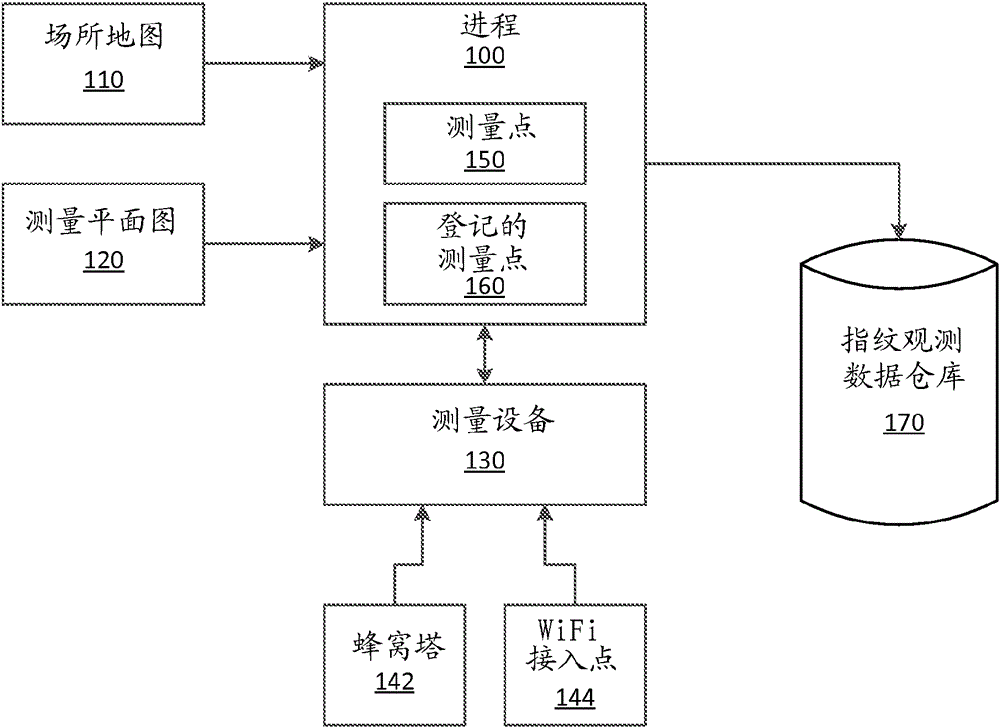
Example apparatus and methods concern rigorous survey-plan based sensor data collection where physical survey locations are correlated to logical locations rather than being tightly coupled to physical map locations. An embodiment includes accessing a venue map and a survey plan associated with the venue map. A survey plan includes a survey path defined by one or more logical survey points. A logical survey point includes a unique co-ordinate free identifier, a description of a recognizable location in the venue, and a co-ordinate configured to register the logical survey point to the corresponding venue map. A surveyor surveys the venue using the survey plan. Surveying the venue includes following the survey plan and acquiring sensor fingerprints at sensor reading points along the survey path. A fingerprint observation data store is populated with survey points that are registered to the survey plan. Survey points include sensor fingerprint data and correlation data.
Owner:MICROSOFT TECH LICENSING LLC
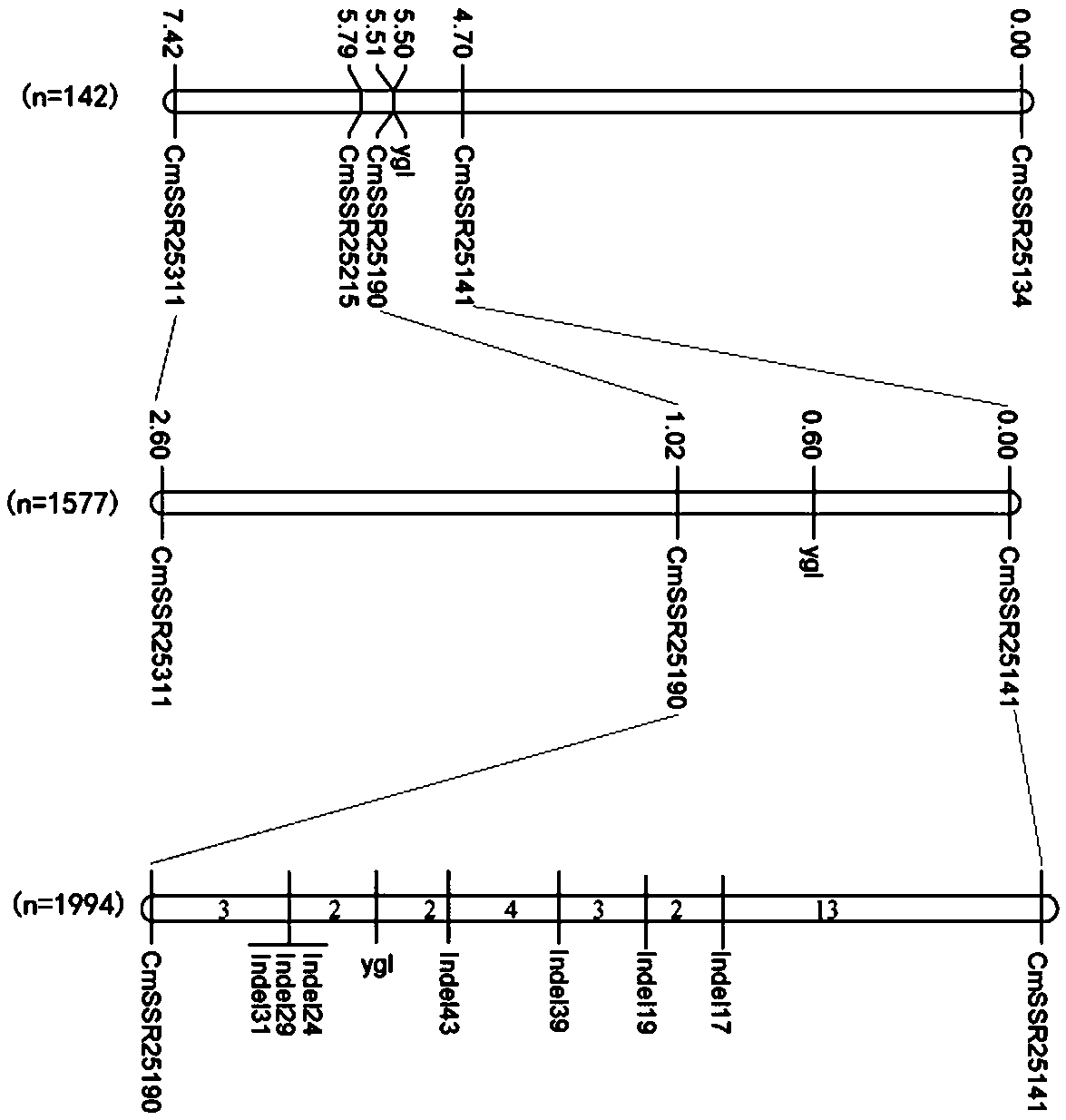
Molecular marker tightly interlocked with muskmelon yellow and green leaf color gene ygl
ActiveCN107868847AImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationAgricultural sciencePhysical Maps
The invention relates to a molecular marker in the technical field of muskmelon genetic engineering, and in particular relates to a pair of molecular markers which are used for positioning a muskmelonyellow and green leaf color gene ygl, and are tightly interlocked with the muskmelon yellow and green leaf color gene ygl. When the molecular marker Indel24 is used for positioning the yellow and green leaf color gene ygl, PCR (Polymerase Chain Reaction) amplification can be carried to obtain a characteristic strip which is tightly interlocked with 191 bp; when the molecular marker Indel43 is used for positioning the yellow and green leaf color gene ygl, PCR amplification can be carried to obtain a characteristic strip which is tightly interlocked with 200 bp. The yellow and green leaf colorgene ygl is located between the two molecular markers, and a physical distance is only about 26Kb. By utilizing the result, a physical map between the two molecular markers can be obtained, so that afoundation is laid for final cloning of the gene ygl and establishment of a molecular marker assisted breeding system; meanwhile, a foundation is also laid for development of muskmelon chloroplasts and researches of a chlorophyll synthesis regulation and control network.
Owner:HENAN AGRICULTURAL UNIVERSITY
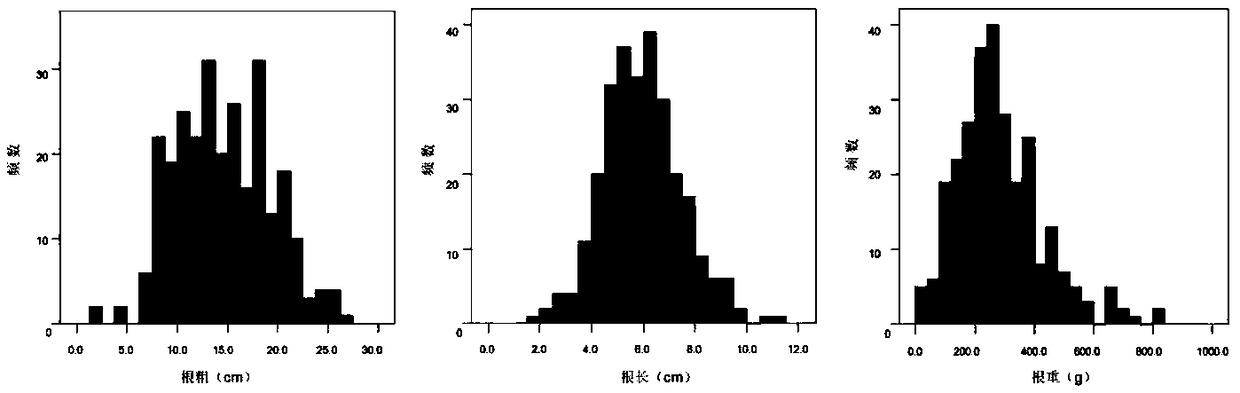
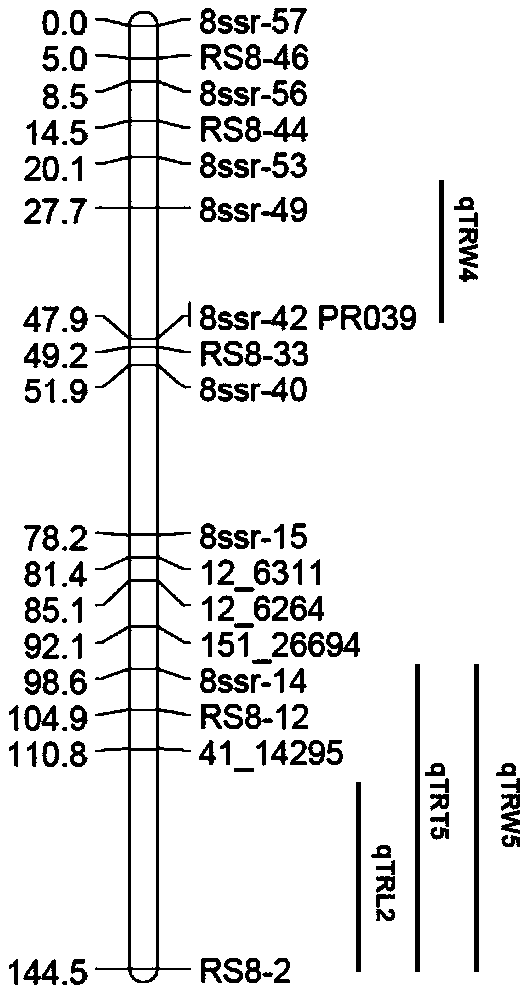
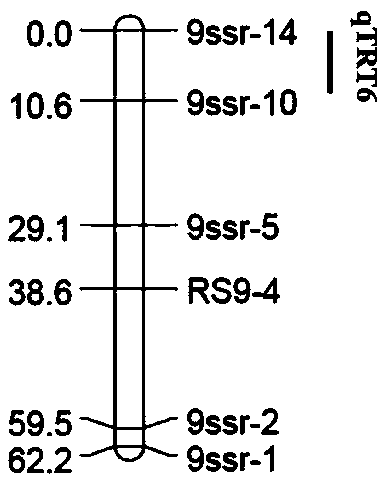
QTLs (quantitative trait loci) of related traits of radish fleshy root and their positioning method
ActiveCN109136400AImprove breeding efficiencyMicrobiological testing/measurementRoot weightStatistical analysis
The invention discloses 13 QTLs (quantitative trait loci) of related traits of radish fleshy root, which locate at chromosomes Chr1, Chr2, Chr3, Chr4, Chr5, Chr6, Chr7 and Chr8 respectively and have gene sequences shown as SEQ ID NO. 1 to SEQ ID NO. 48, and also discloses a method of positioning the above QTLs. The method includes: (1) constructing F2 segregation population; (2) acquiring and statistically analyzing related phenotypic trait data (root length, root thickness and root weight) of fleshy root of female and male parents and F1 and F2 segregation populations; (3) extracting DNA forthe female and male parents and F1 and F2 segregation populations; (4) screening polymorphic SSR (simple sequence repeat) primers, and genotyping the female and male parents and F1 and F2 segregationpopulations; (5) constructing a genetic map; (6) using the software MapQTL 4.0 to perform QTLs analysis. The QTLs and the method have the advantages that linkage map of radishes is connected to its genome physical map for the first time, 2 QTLs to control the radish root length are positioned for the first time, and variety breeding efficiency is improved accordingly.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
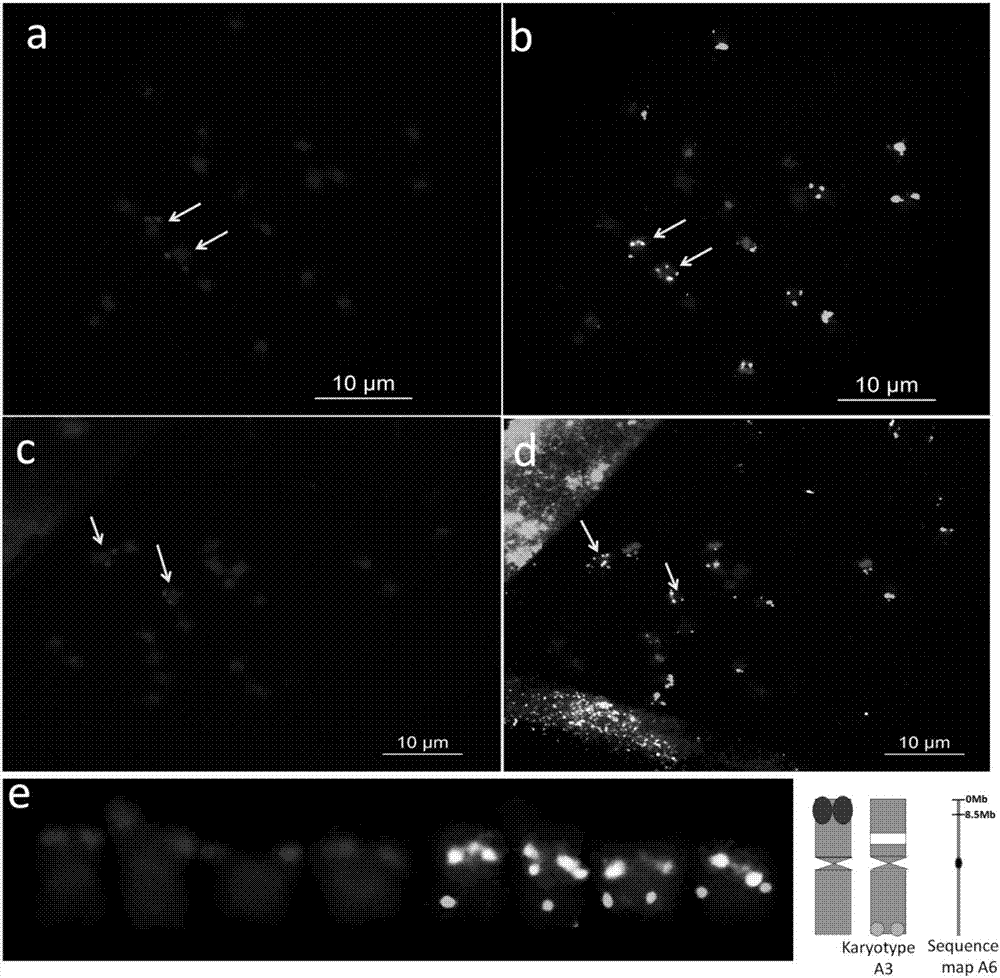
Method for corresponding peanut genome chromosome sequence map to actual karyotype chromosome number
ActiveCN107130033AConvenient researchEfficient use ofMicrobiological testing/measurementWild speciesIn situ hybridisation
The present invention discloses a method for corresponding a peanut genome chromosome sequence map to an actual karyotype chromosome number. According to the method, a peanut chromosome single copy oligonucleotide library is developed by using a peanut wild species A.duranensis genome sequence sketch, and the correspondence between the peanut genome chromosome sequence map and the actual karyotype chromosome number is achieved through probe labeling and sequential fluorescence in situ hybridization. According to the present invention, the correspondence between the sequence map of the A6 chromosome of the peanut wild species A.duranensis genome and the chromosome having the actual karyotype chromosome number of A3 is firstly achieved, such that the foundation is established for the integration of peanut genome information, chromosome labeling information and genetic linkage map information so as to develop the new approach for the revealing of the peanut chromosome exchange, recombination and evolution law; and the method can be used for the development of peanut breeding research, can establish the foundation for the construction of the peanut physical map and the physical locating of the single gene, and can promote the research and utilization of the gene.
Owner:HENAN ACAD OF AGRI SCI
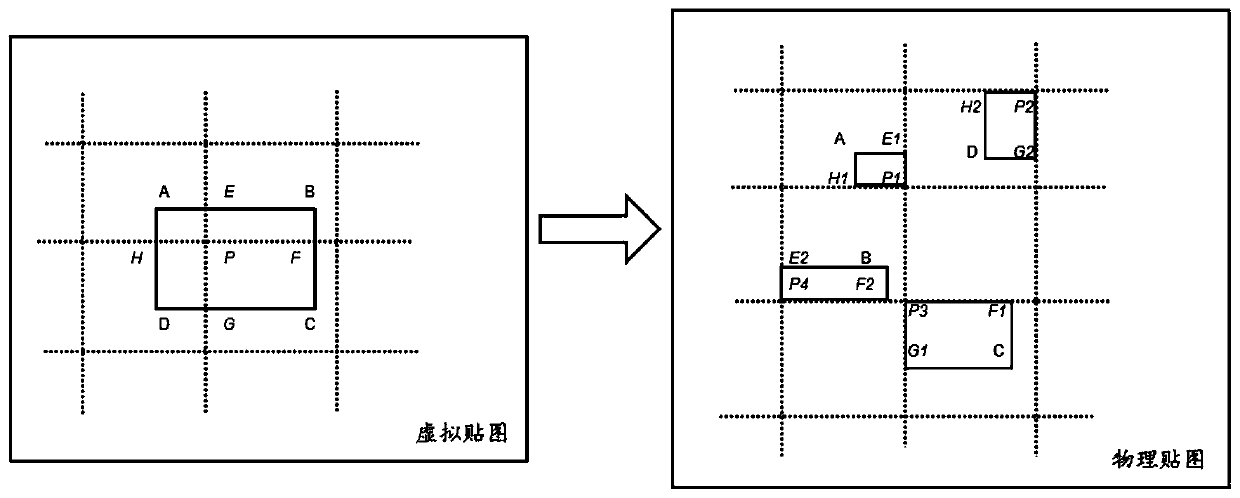
Map processing method and device, computer readable storage medium and electronic device
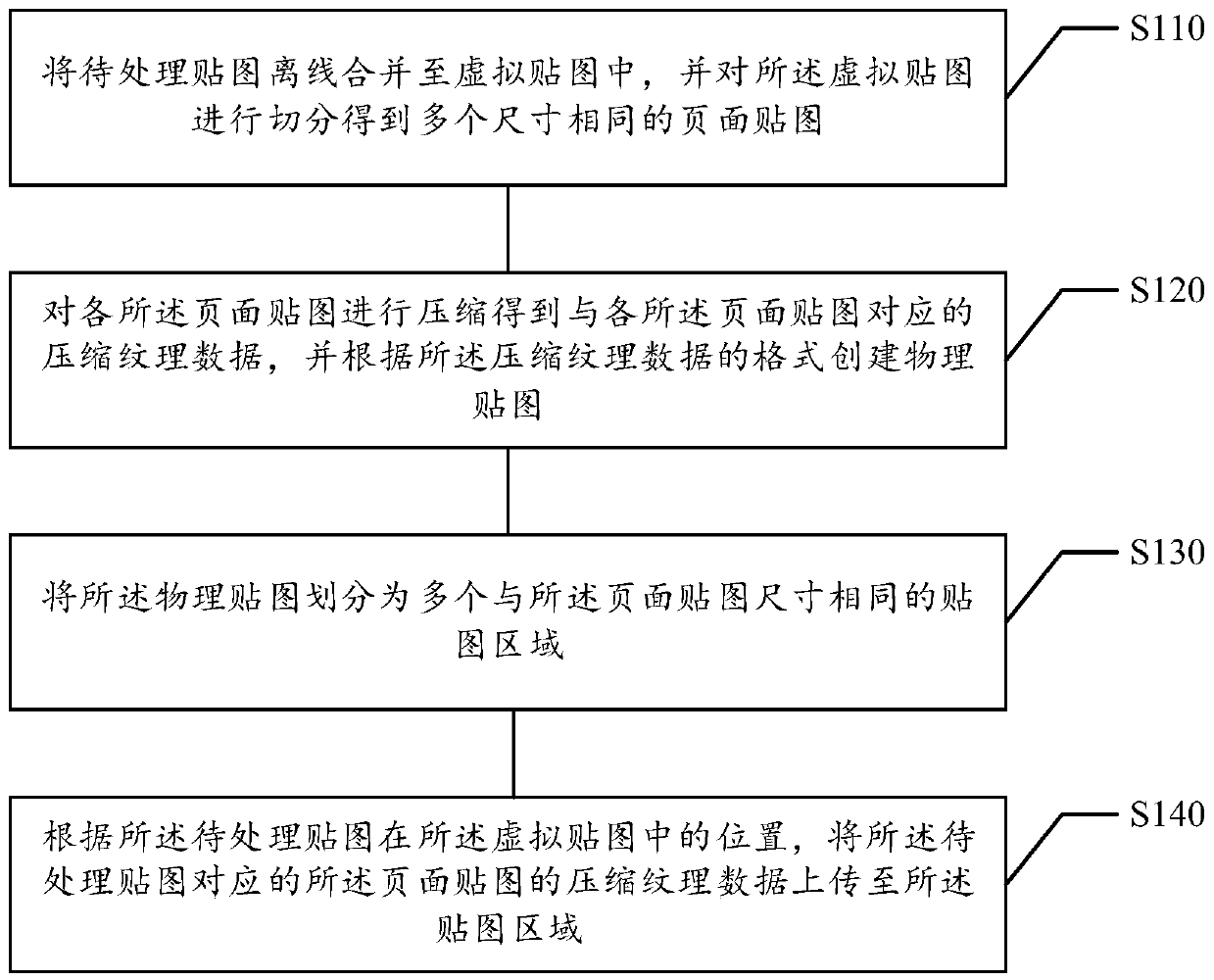
ActiveCN110609726AExecution for user interfacesEnergy efficient computingOccupancy rateImaging processing
The embodiment of the invention relates to a map processing method and device, a computer readable storage medium and an electronic device, and relates to the technical field of image processing. Themethod comprises the steps of enabling a to-be-processed map to be merged into a virtual map offline, and segmenting the virtual map to obtain a plurality of page maps with the same size; compressingeach page map to obtain the compressed texture data corresponding to each page map, and creating a physical map according to the format of the compressed texture data; dividing the physical map into aplurality of map areas with the same size as the page map; and uploading the compressed texture data of the page map corresponding to the to-be-processed map to the map area according to the positionof the to-be-processed map in the virtual map. According to the embodiment of the invention, the utilization rate of the virtual map of the maps is improved while the memory occupancy rate is reduced.
Owner:NETEASE (HANGZHOU) NETWORK CO LTD
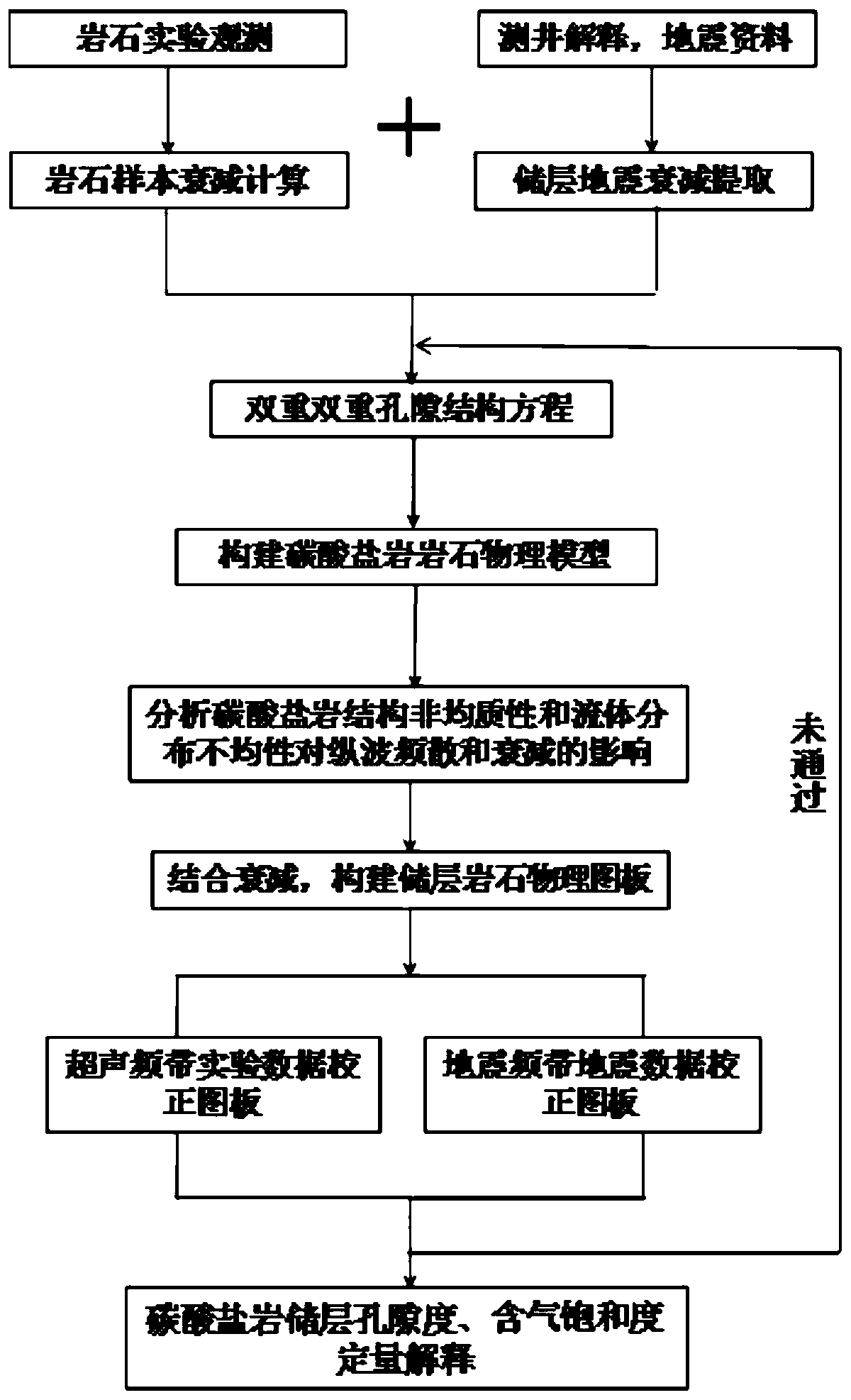
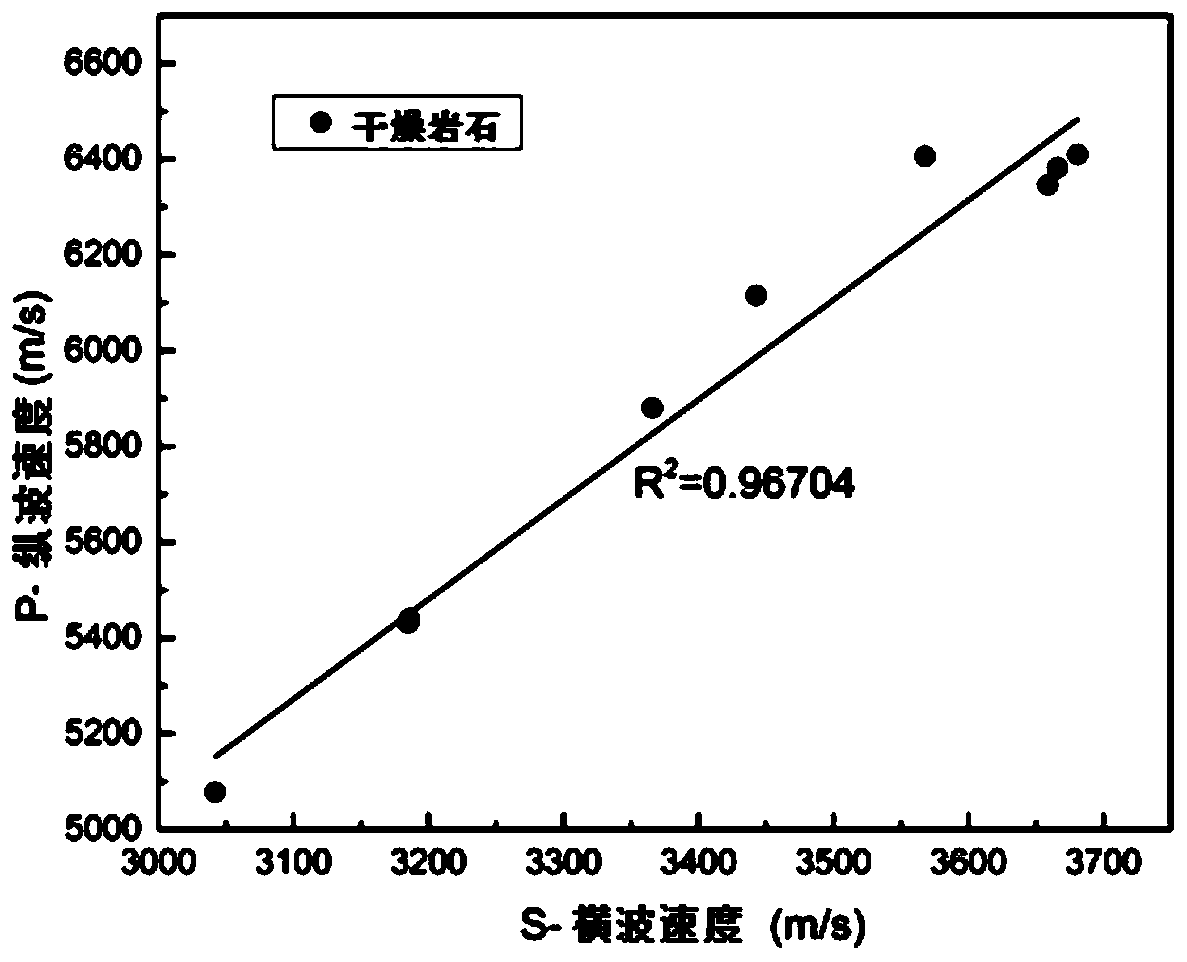
Method for identifying fluid saturation of carbonate reservoir based on poststack seismic data
ActiveCN110456412AStrong and stable structureIncreased sensitivitySeismic signal processingExperimental testingUltrasound attenuation
The invention discloses a method for identifying the fluid saturation of the carbonate reservoir based on poststack seismic data. The method comprises following steps: step one, carrying out ultrasonic experimental testing of carbonate reservoir rocks and calculation of rock sample attenuation; step two, carrying out seismic attenuation extraction of the carbonate reservoir; step three, constructing a carbonatite attenuation rock attenuation physical model; step four, analyzing influences on the dispersion and attenuation by anisotropy and fluid distribution heterogeneity of the carbonatite structure; step five, constructing a carbonatite attenuation rock physical map plate; step six, correcting the attenuation rock physical map plate under the ultrasonic and seismic frequency bands; and step seven, carrying out quantitative interpretation of the carbonate reservoir porosity and fluid saturation. During the ultrasonic experimental testing, ultrasonic waveform measurement under the partially saturated condition is carried out on the carbonate reservoir sample and the gas is nitrogen. According to the invention, influences on the elastic wave dispersion and attenuation by the anisotropy of the pore structure and the uneven distribution of the partially saturated fluid in the complex rock can be described based on the dual-dual pore structure model.
Owner:HOHAI UNIV
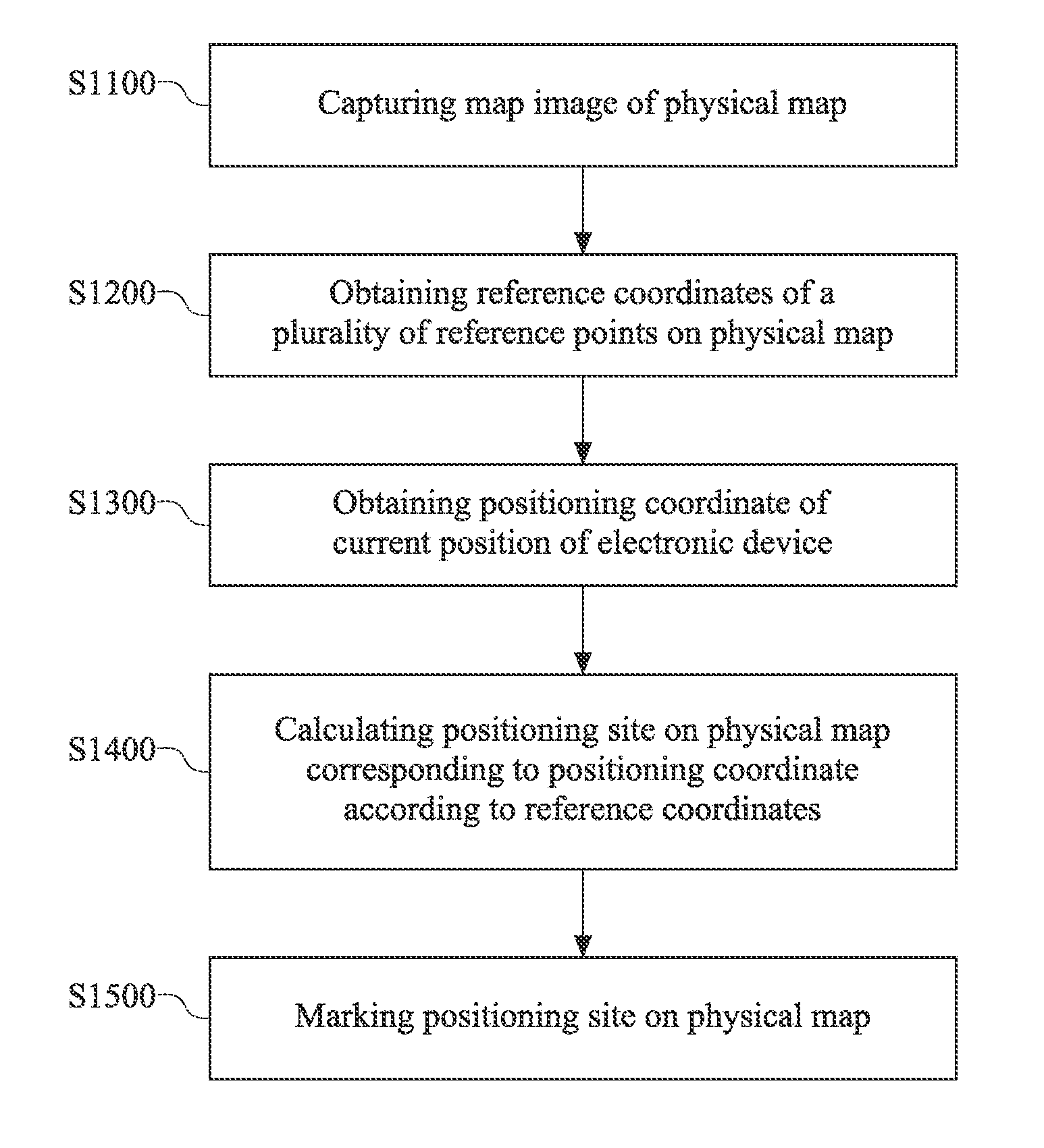
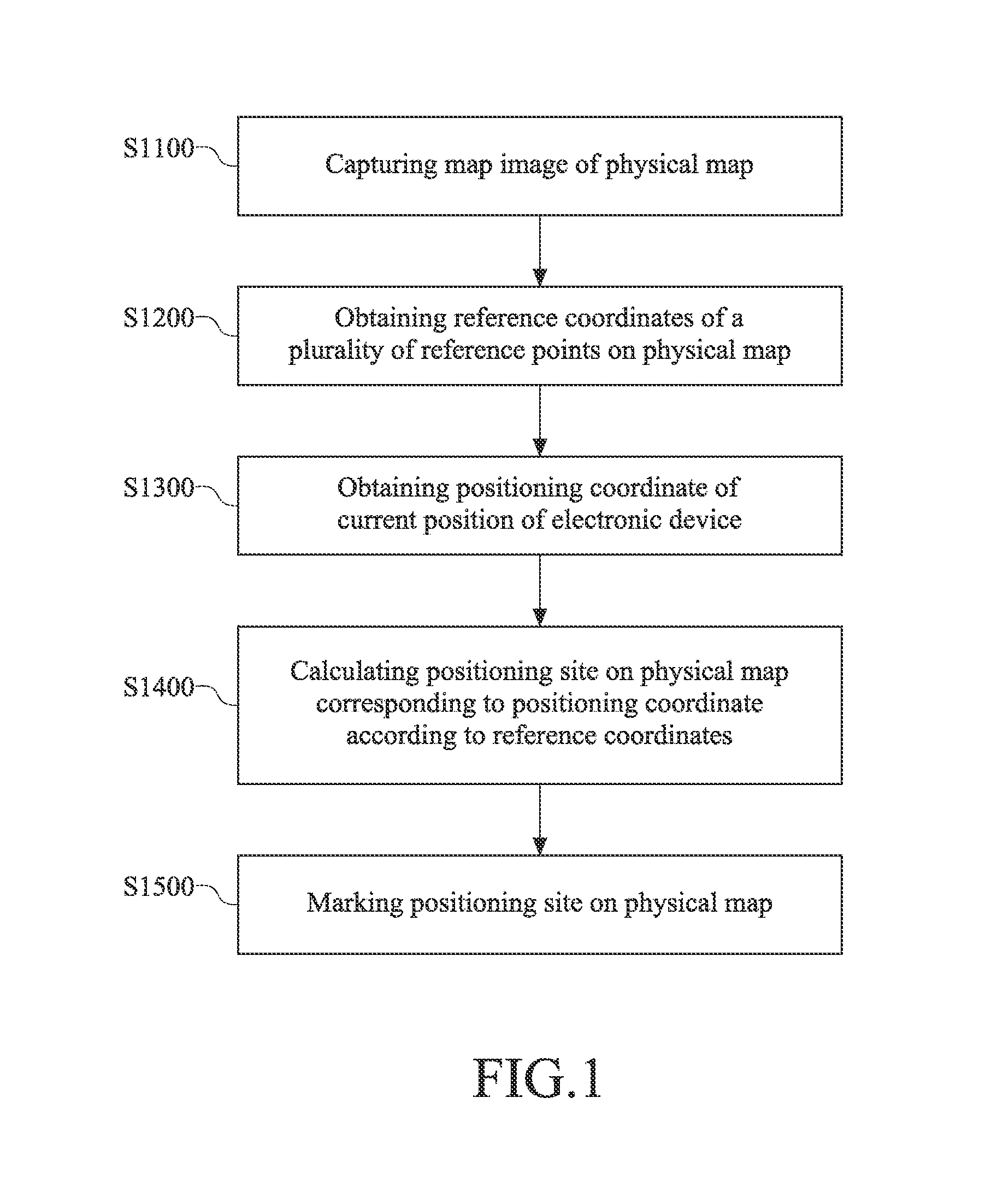
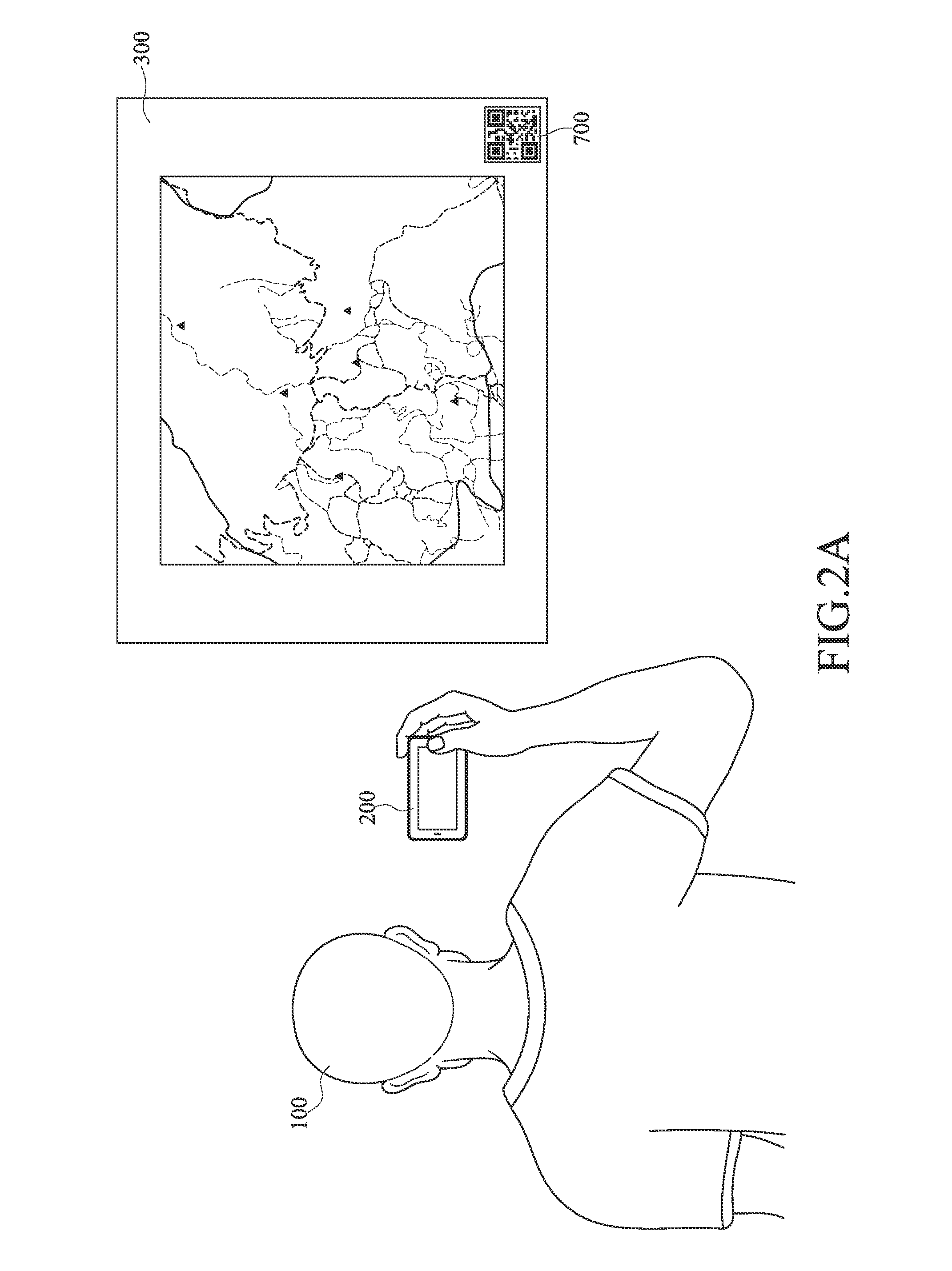
Map marking method
InactiveUS20160153784A1Navigational calculation instrumentsRoad vehicles traffic controlPhysical MapsGlobal Positioning System
A map marking method comprises capturing a map image from a physical map, capturing a plurality of reference coordinates corresponding to the physical map, obtaining a positioning coordinate from the global positioning system, calculating a positioning site on the physical map corresponding to the positioning coordinate according to the plurality of reference coordinates, the map image, and the positioning coordinate, and marking the positioning site on the physical map. As such, a user may arbitrarily mark his / her own position on a physical map with his / her positioning device utilizing the above mentioned method, and may explore the world more conveniently.
Owner:INVENTEC PUDONG TECH CORPOARTION +1
Brachypodium distachyon functional centromere antigen polypeptide and application
InactiveCN105461789ADiscuss formationDiscuss functionSerum immunoglobulinsImmunoglobulins against plantsNew Zealand white rabbitNucleotide
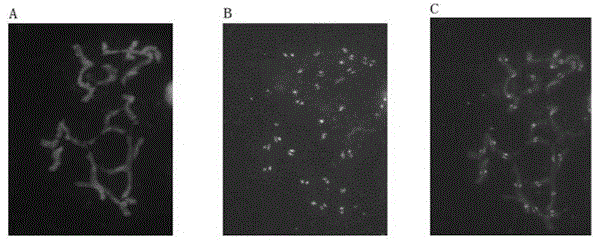
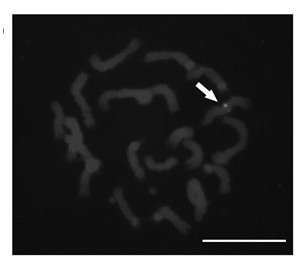
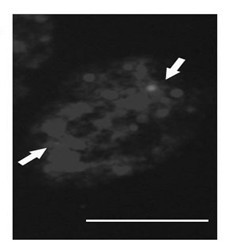
The invention provides a brachypodium distachyon functional centromere histone CENH3 and application. Firstly, the nucleotide sequence of a brachypodium distachyon functional centromere histone CENH3 gene is predicted through the bioinformatics sequence comparison method, a polypeptide is synthesized according to a segment of specific sequence of the N end of an amino acid sequence encoded by the nucleotide sequence, New Zealand white rabbits are conventionally immuned through the polypeptide, antiserums are prepared, and a CENH3 protein antibody is obtained through antigen affinity purification. The CENH3 antibody can be used for immunostaining experiments, physical positioning of centromere and chromatin immunoprecipitation (ChIP) experiments, the functional DNA sequence of a brachypodium distachyon functional centromere is obtained, positioning of centromere genome genetic maps and physical maps is carried out, and important tools and information are provided for research of centromere formation and function exercise mechanisms and building of artificial chromosomes.
Owner:FUJIAN AGRI & FORESTRY UNIV
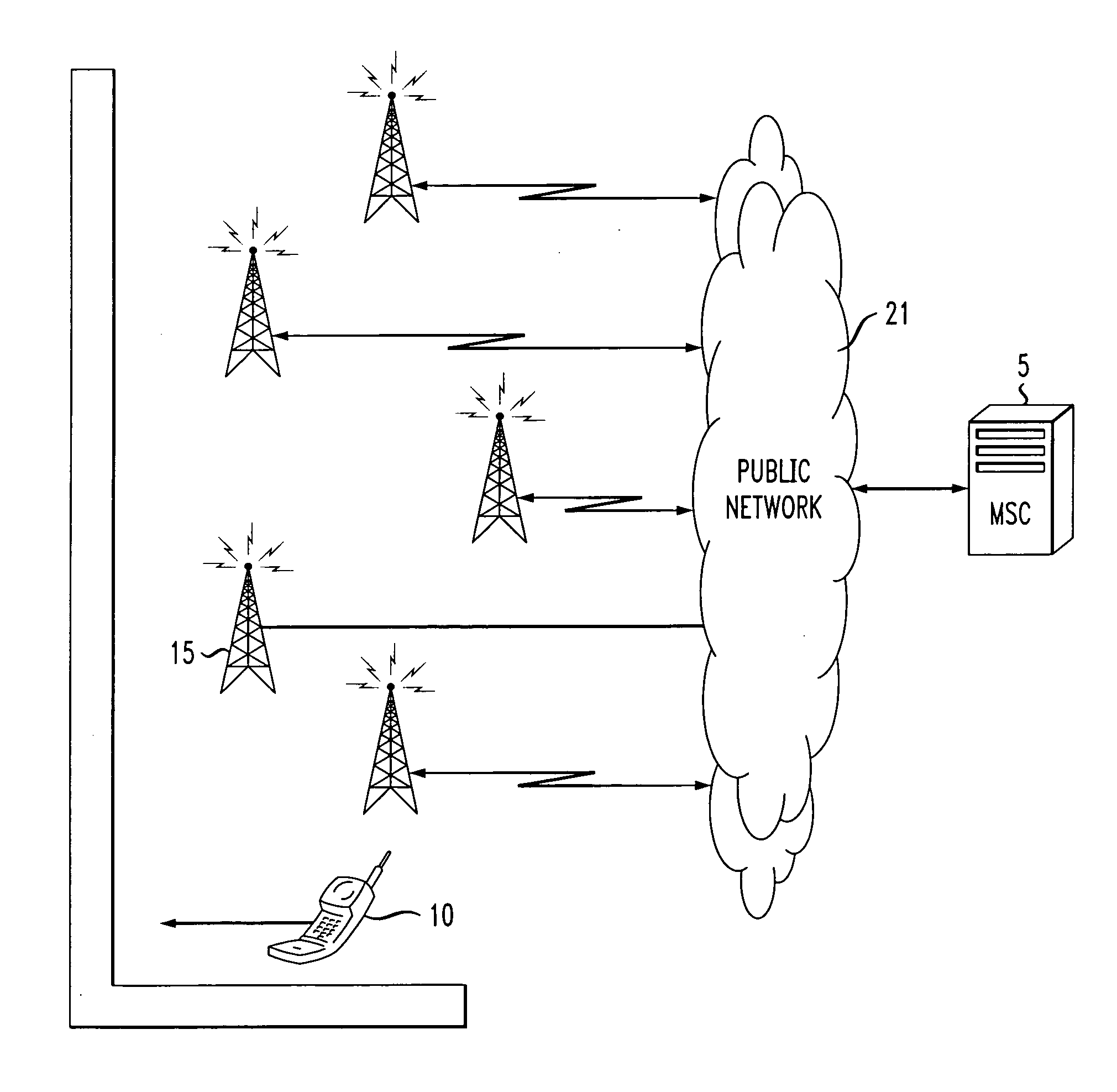
Method and apparatus for automated mapping cell handset location data to physical maps for data mining (traffic patterns, new roads)
InactiveUS20070218872A1Unauthorised/fraudulent call preventionEavesdropping prevention circuitsTraffic countPhysical Maps
A method and system for the automated mapping of a mobile station and / or handset are provided. The technique includes capturing the identification information of a mobile station, linking a unique tag to the identification information, releasing the identification information of the mobile station, while maintaining the unique tag, tracking the movement of the mobile station through the use of the unique tag and releasing the unique tag after a predetermined amount of time. This method and system could be used for mapping and data mining of traffic flow patterns. As such, this method and system will be useful in determining the amount of use a road may receive automatically, without having traffic counters. It can also be used in determining automatically when new roads have become opened. Furthermore, this data could be used for foot traffic flow through a building.
Owner:LUCENT TECH INC
Evaluation method of Gossypium hirsutum-Gossypium barbadense introgression lines
ActiveCN106048033AGood for physical locationAssisted selection breedingMicrobiological testing/measurementAgricultural sciencePhysical Maps
The invention relates to an evaluation method of Gossypium hirsutum-Gossypium barbadense introgression lines. The invention discloses a comprehensive evaluation method of Gossypium hirsutum-Gossypium barbadense introgression lines. The method comprises the following steps: (1) identifying verticillium wilt resistance; (2) carrying out molecular marker analysis on Gossypium hirsutum-Gossypium barbadense introgression lines; (3) determining the physical location of the molecular marker in the cotton genome; and (4) according to the physical location of the molecular marker, carrying out physical location on the introduced disease-resistant chromosome segment, and drawing a physical map of the Gossypium hirsutum-Gossypium barbadense introgression lines. By combining the verticillium wilt resistance identification and molecular marker analysis, the molecular marker sequence is compared with the tetraploid Gossypium hirsutum genome, and the disease-resistant gene carried in the Gossypium hirsutum-Gossypium barbadense introgression lines is located, thereby laying the foundation for disease-resistant gene cloning and disease-resistant variety improvement.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI
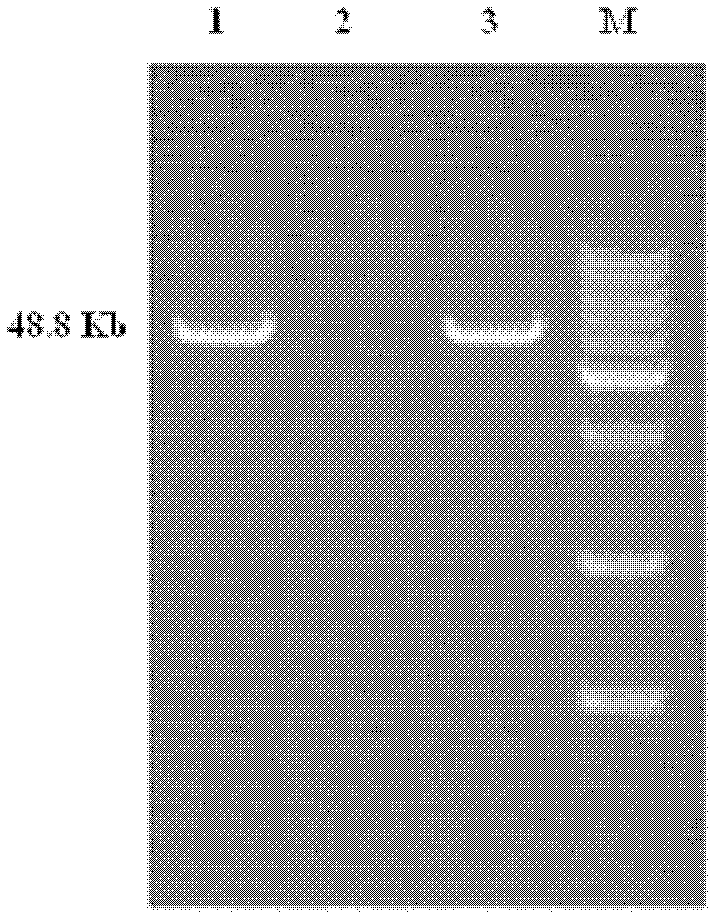
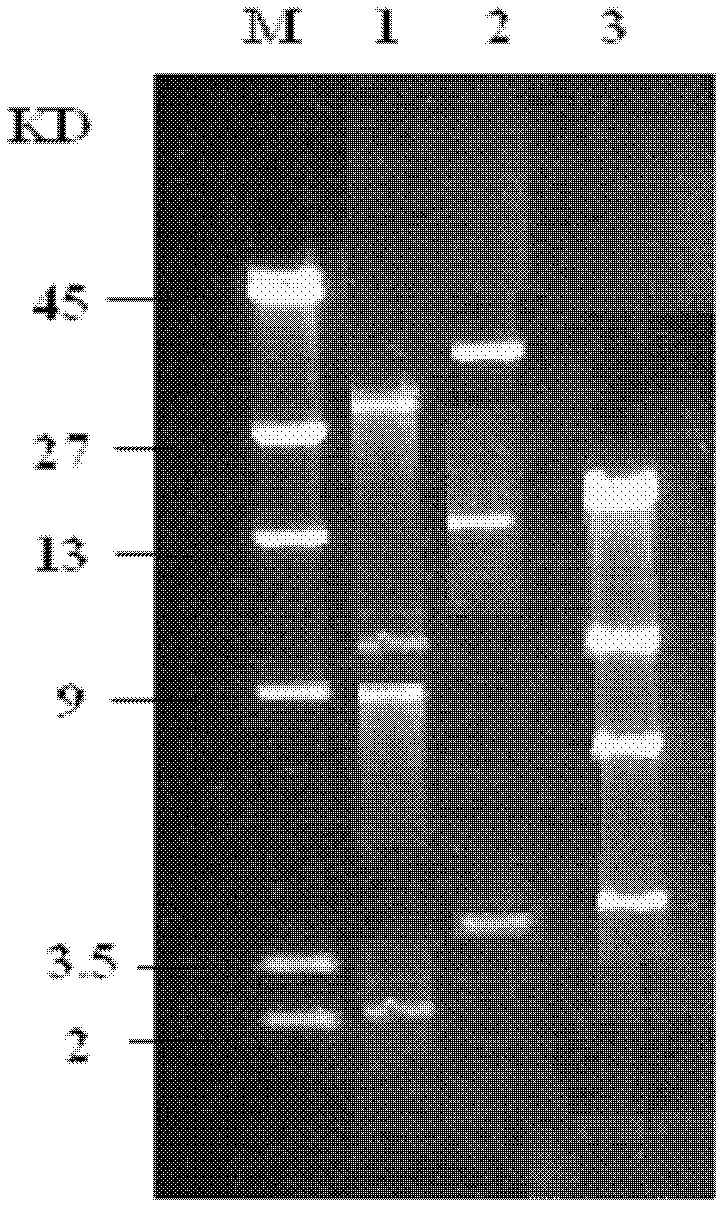
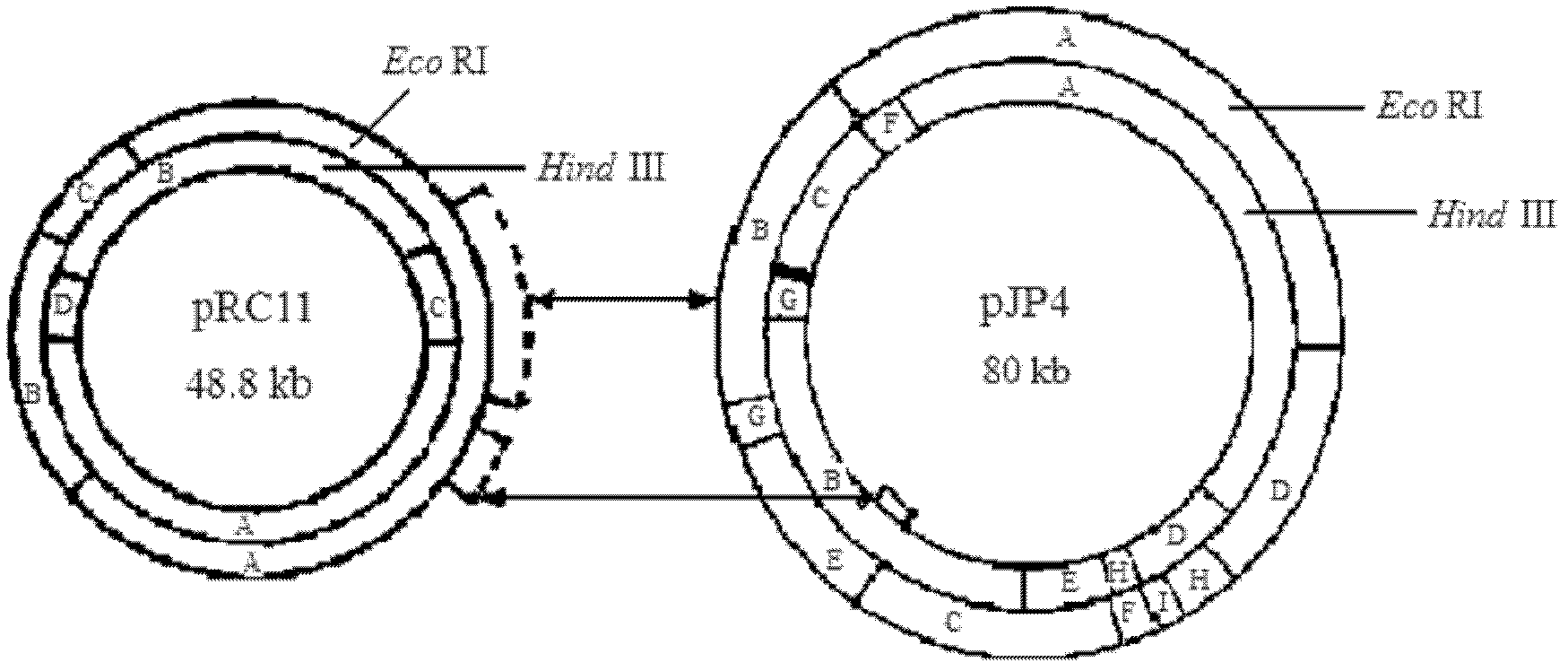
Chlorinated aliphatic hydrocarbon degradative plasmid pRC11, engineered bacteria and application thereof
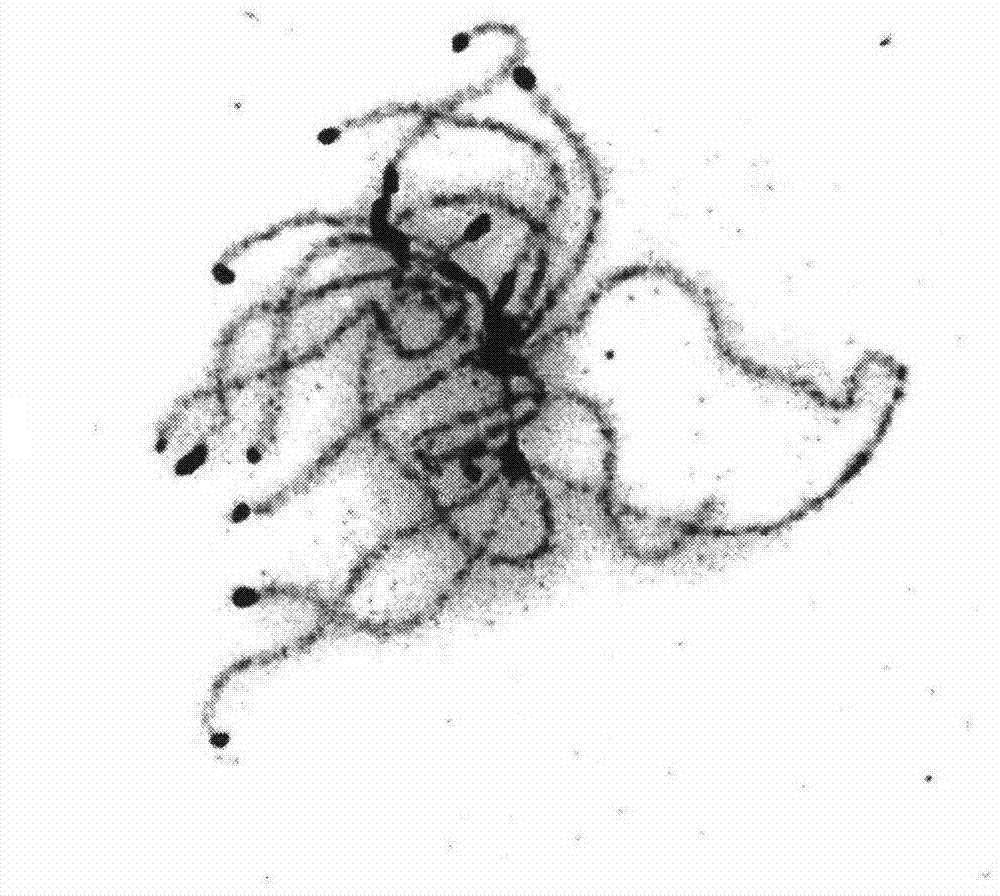
InactiveCN102321659AEliminate pollutionClear genetic backgroundBacteriaDispersed particle separationPhysical MapsEthyl Chloride
The invention provides a chlorinated aliphatic hydrocarbon degradative plasmid pRC11, the physical map of which is shown in Figure 3, and the nucleotide sequence of which is shown as SEQ ID No.1. The plasmid provided by the invention is obtained through screening from an industrial waste gas biological purification microbial resource strain library self-built by a laboratory. The beneficial effects of the invention are mainly reflected in that: the chlorinated aliphatic hydrocarbon degradative plasmid - pRC11 is provided, and the physical map of the plasmid is built; the genetic background of the plasmid is very clear, and the plasmid can be used for molecular breeding, and nurturing efficient multi-functional plasmid bacteria; and the biodegradation effect of halogenated hydrocarbon organic pollutants taking methylene chloride as a representative is improved, and the processing capacity of organic waste gas is substantially improved.
Owner:ZHEJIANG UNIV OF TECH
Detection method of vegetal single copy genes
InactiveCN102191325AThe detection process is shortHigh sensitivityMicrobiological testing/measurementFluorescence/phosphorescencePhysical MapsSingle copy
The invention relates to a detection method of vegetal single copy genes based on fluorescence in situ hybridization of quantum dots. By preparing quantum dot coupled DNA (deoxyribonucleic acid) as a probe, according to the traditional fluorescence in situ hybridization method, the hybridization detection of vegetal single copy genes is carried out on interphase nucleuses, chromosomes, DNA fibersor the like. In the method provided by the invention, a direct marking method is adopted, thus an antibody detection process is omitted; and because the quantum dots have high fluorescence intensity and can resist photo-bleaching, the sensitivity and resolution of hybridization are improved, and the vegetal single copy genes can be quickly and effectively located. The method provided by the invention can be widely used for drawing physical maps and analyzing the composition, structure, rearrangement, evolutionary relationship and the like of genomes.
Owner:WUHAN UNIV
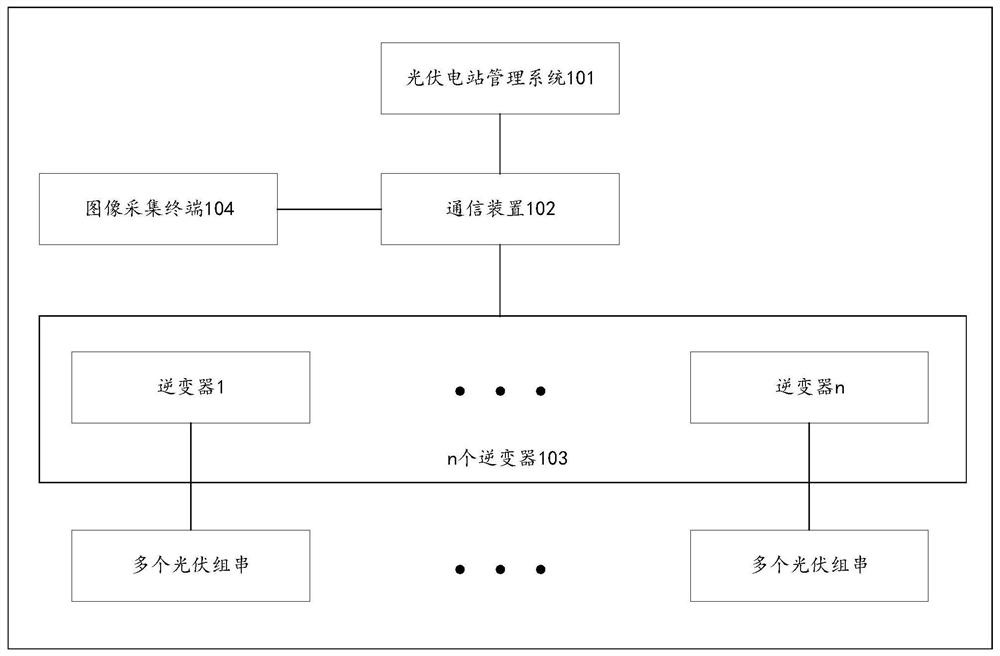
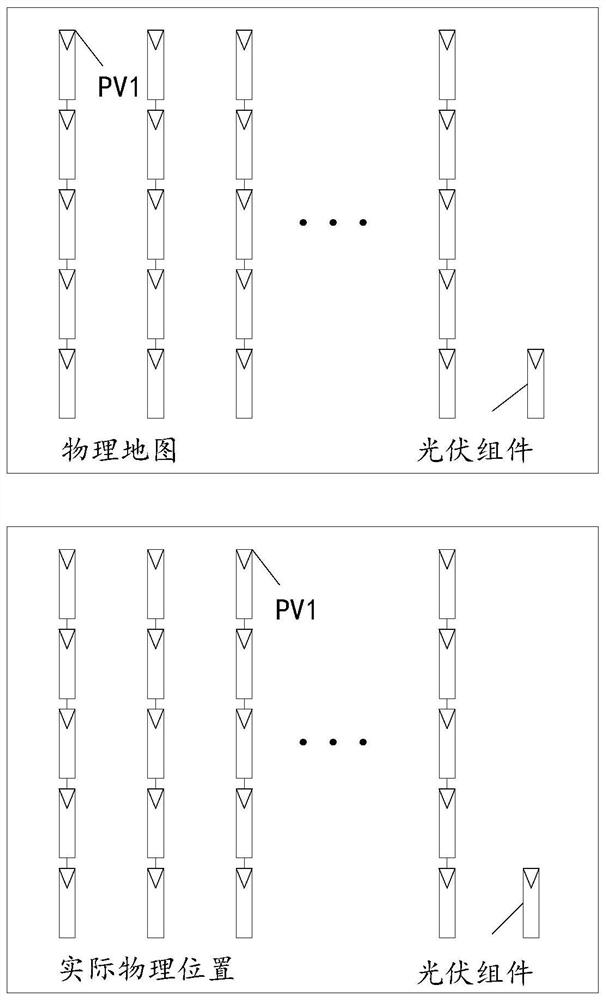
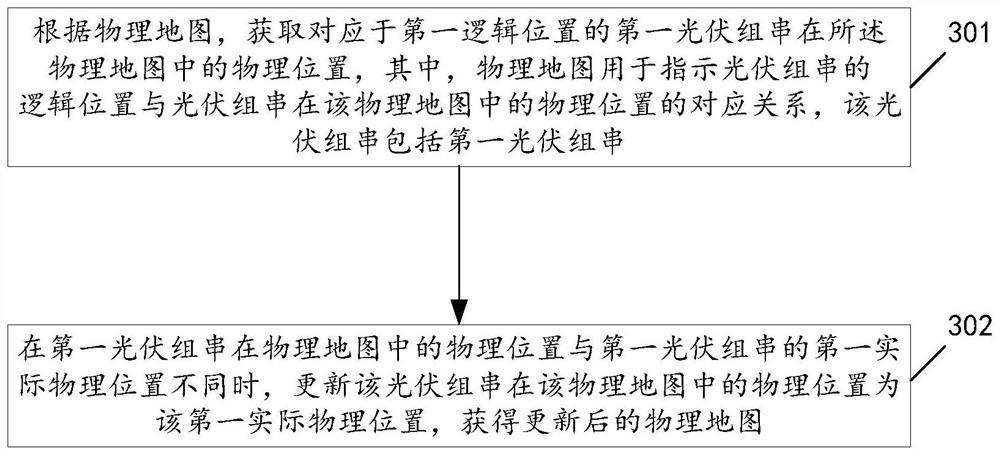
Photovoltaic string position updating method and device
PendingCN113157830AGeneration forecast in ac networkDatabase updatingComputational physicsPhysical Maps
Owner:HUAWEI DIGITAL POWER TECH CO LTD
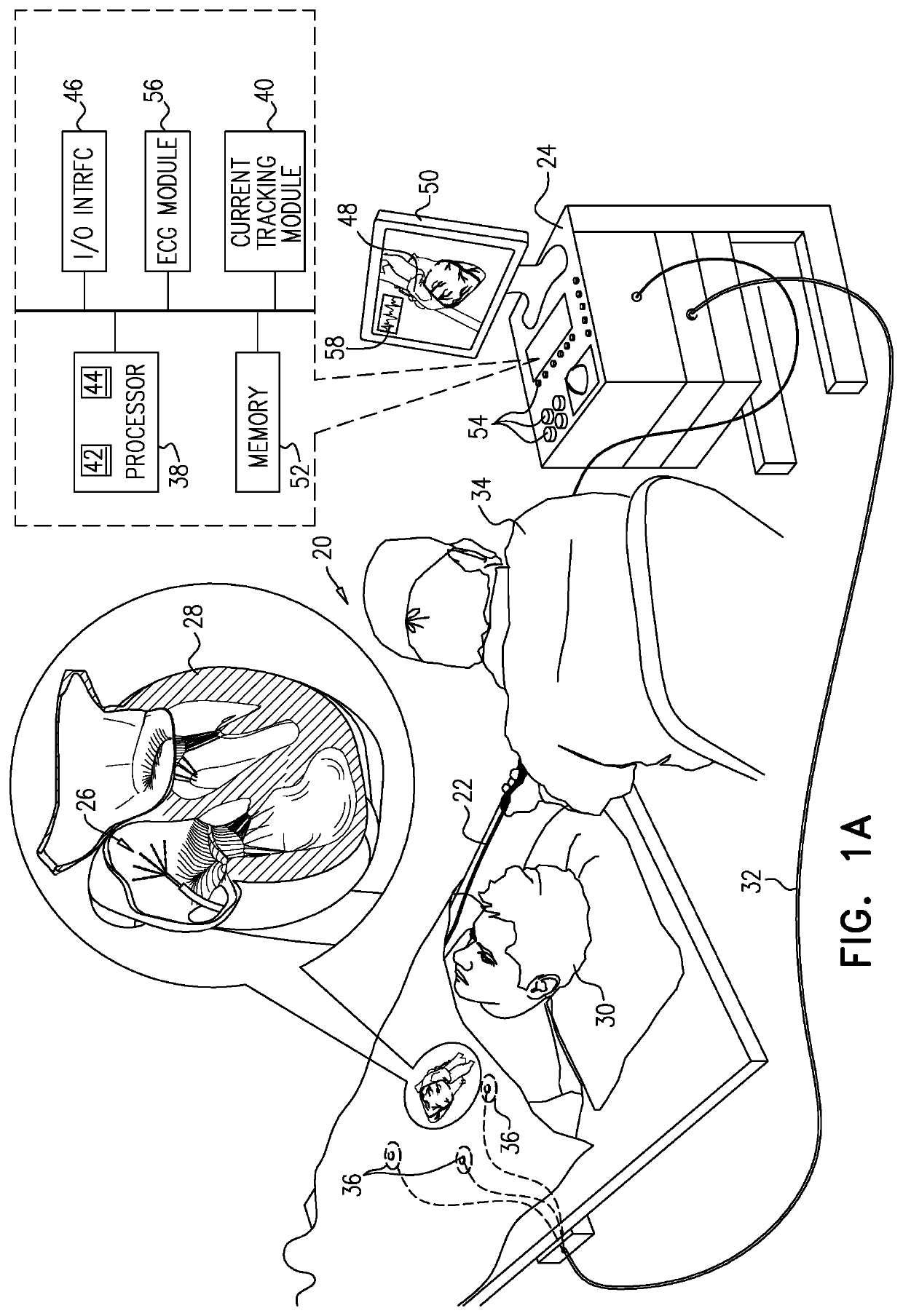
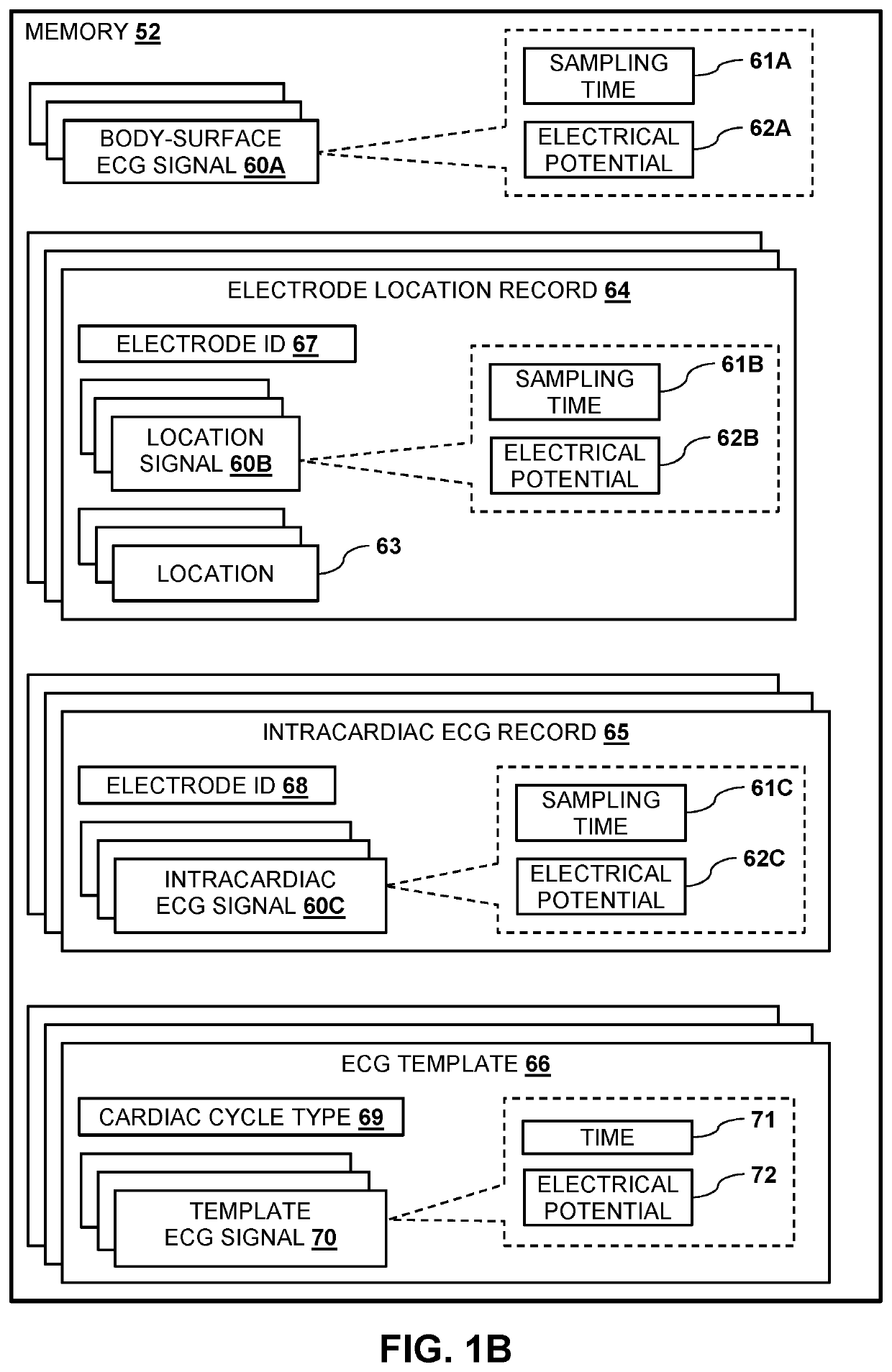
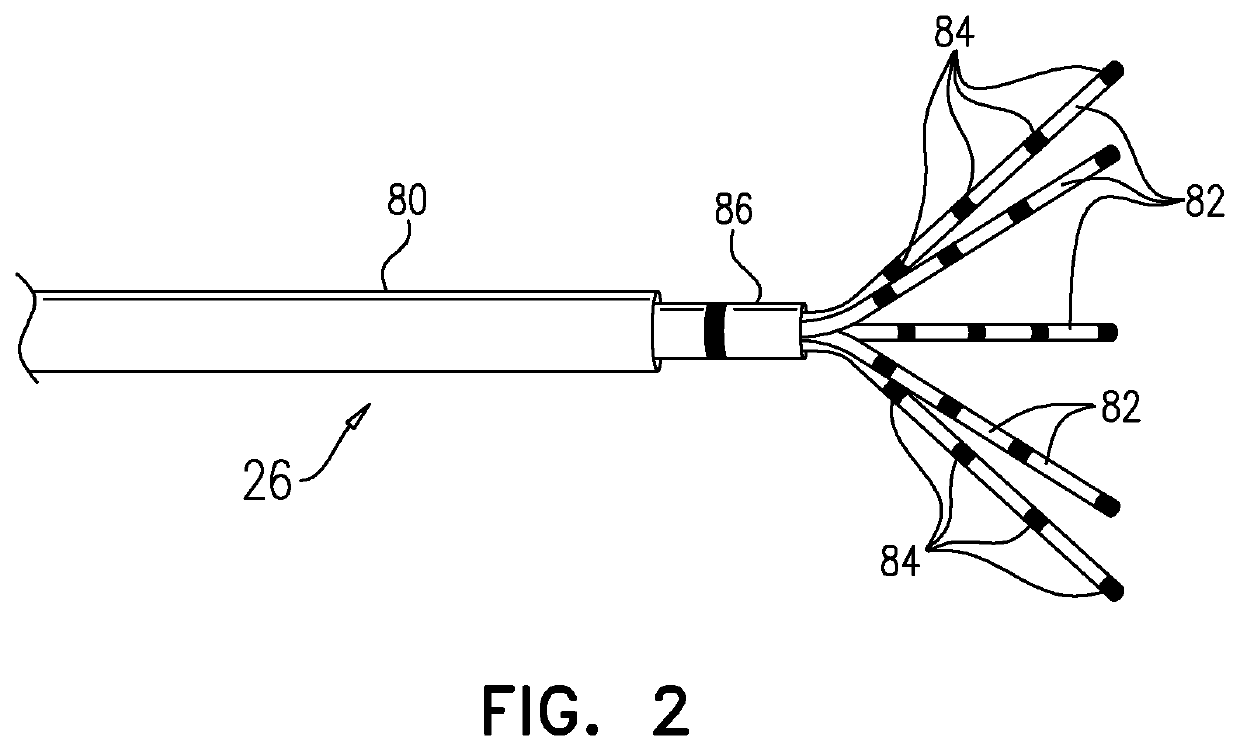
Mapping local activation times for sinus and non-sinus cardiac cycles
A method, including receiving sets of signals during multiple cardiac cycles, each set indicating, for a probe inserted into a cardiac chamber, a 3D location of a distal end of the probe, electrical potentials measured at the location, and respective times during a given cycle when the potentials were measured. The received measurements and the respective times are compared to a first template for a sinus rhythm cycle and a second template for a non-sinus rhythm cycle so as to identify a sequence of cycles including consecutive first, second, and third cycles wherein the first and second cycles match the first template and the third cycle matches the second template. A physical map is generated based on the locations. Based on the received locations and corresponding potentials, an electroanatomic map including the local activation times for the non-sinus rhythm cycle overlaid on the physical map is rendered to a display.
Owner:BIOSENSE WEBSTER (ISRAEL) LTD
Restriction enzyme based whole genome sequencing
InactiveUS8932812B2Maximize possibilitySugar derivativesMicrobiological testing/measurementWhole genome sequencingA-DNA
Owner:KEYGENE NV
Stapler with adapter
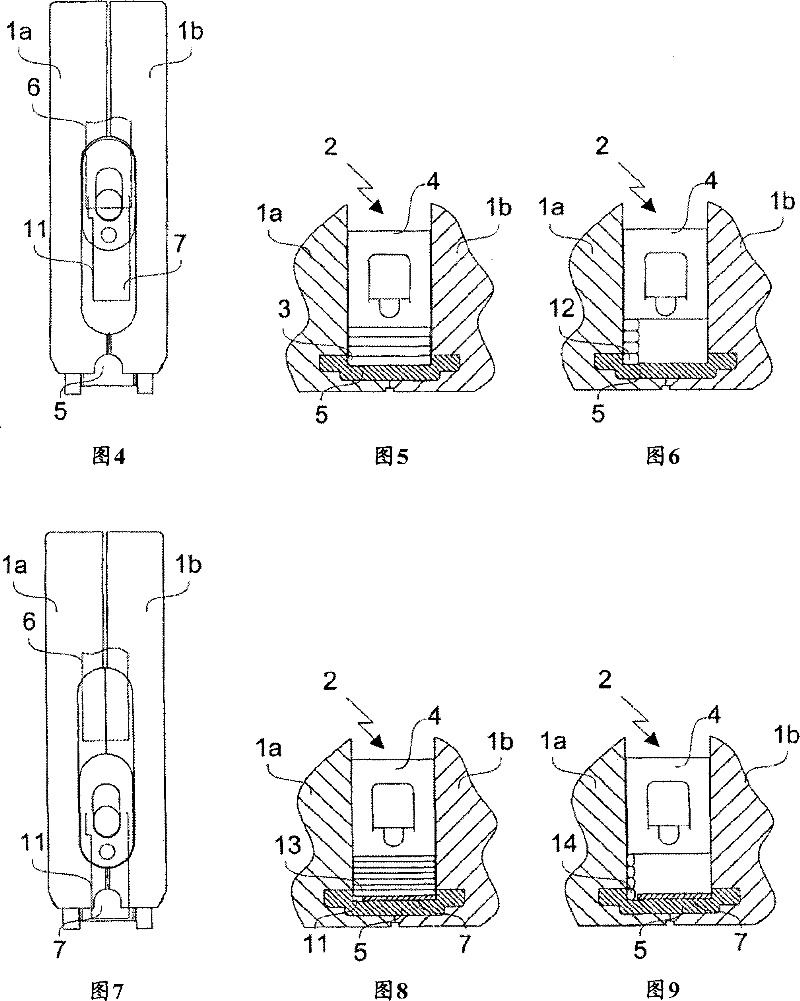
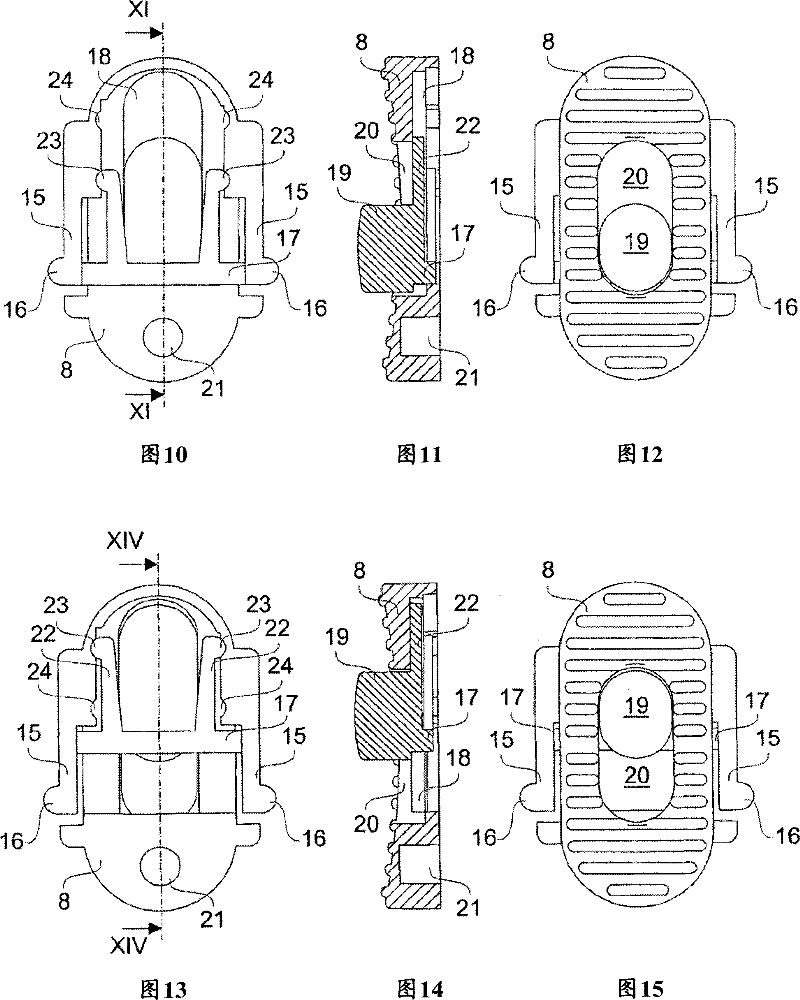
InactiveCN101076434BAvoid accidental slidingSimple structureStapling toolsPhysical MapsBase sequence
To provide a method and an apparatus for decoding genome base sequences in which whole genome base sequences can be restored by recovering DNA from a chromosome while recording the presence position and determining the base sequences, and a method and an apparatus for preparing a genome physical map in which positioning of a genome library clone onto a genome is carried out by recovering DNA froma specific position on the chromosome and utilizing a part of the base sequence information. The shape of the chromosome is precisely acquired by an AFM search needle 7 also having a scraping mechanism, and minute fragments at accurate positions on the chromosome are continuously scraped. The scraped chromosome fragments are charged to an extraction means 8 of DNA and the extracted DNA is transferred to an amplification means 9. The amplified DNA is cloned to an adequate vector and a base sequence of a clone is successively determined by a base sequence-determining means 10. The resultant base sequence data are fed to a data processing mechanism 11, and the base sequences of the fragments and the whole chromosomes are restored.
Owner:ROMEO MAESTRI & FIGLI
Fluorescence in situ hybridization method for high resolution chromosomes of cucumber
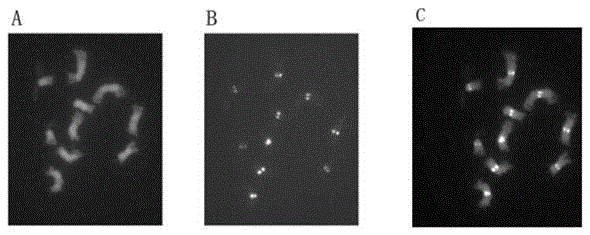
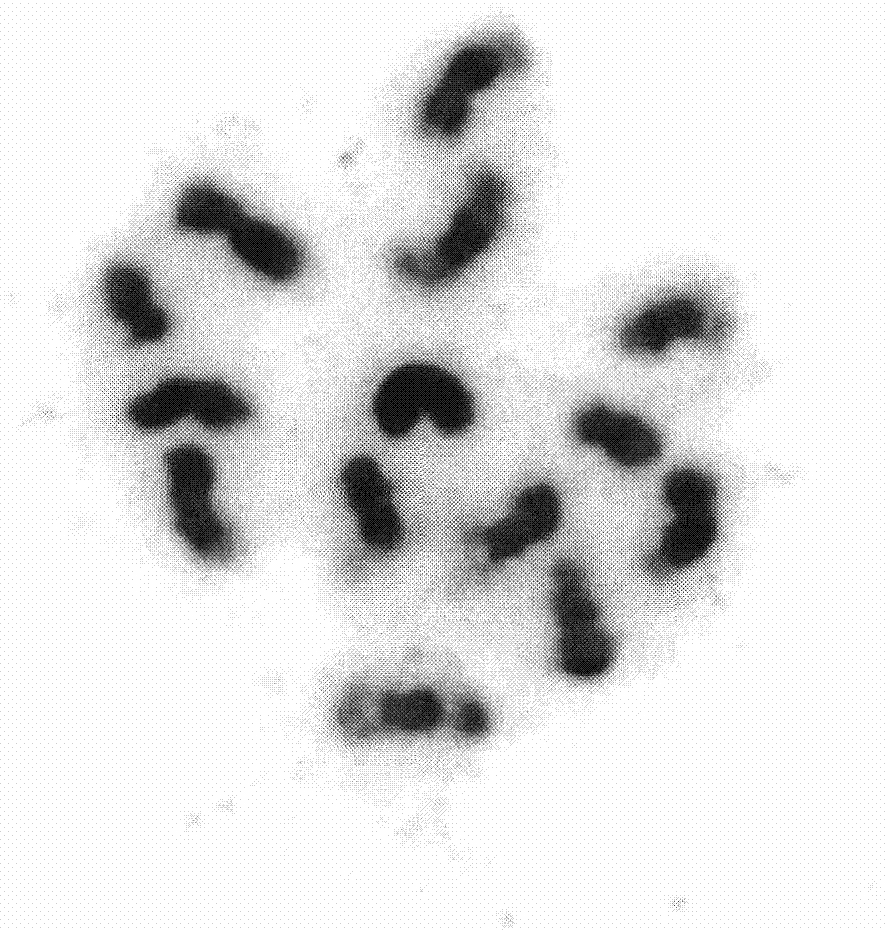
InactiveCN103114151AConducive to carrying out cytological researchImprove the recognition rateMicrobiological testing/measurementFluorescence/phosphorescenceTarget signalFluorescence
The invention provides a fluorescence in situ hybridization method, wherein fluorescence in situ hybridization with multiple probes is performed based on chromosomes in the meiosis pachytene stage of cucumber as a target DNA (Deoxyribonucleic Acid). The method comprises the following steps of: selecting high quality films with good chromosome dispersion and clear morphological structure, degenerating in 70% deionized formamide at 80 DEG C, and dehydrating and drying in gradient alcohol; extracting a target plasmid DNA by alkaline lysis, marking by an incision translation method, preparing a hybrid mixed liquor, degenerating at 90 DEG C; and loading to a glass slide to hybridize the target DNA and probe; and finally detecting signals. The fluorescence in situ hybridization technology provided by the invention can be applied to drawing a physical map of chromosomes of cucumber. As multiple probes can be added at one time, repeated hybridization to one film can be avoided, so that the experimental efficiency is greatly improved. According to the invention, appropriate whole genome DNA and Cot-1DNA of cucumber are added in the process of configuring the hybrid mixed liquor for blocking spurious signals and repetitive sequence signals, so that the identification rate of target signals is improved and the accuracy of experimental result is increased.
Owner:NANJING AGRICULTURAL UNIVERSITY
Detection method of vegetal single copy genes
InactiveCN102191325BThe detection process is shortHigh sensitivityMicrobiological testing/measurementPhysical MapsSingle copy
The invention relates to a detection method of vegetal single copy genes based on fluorescence in situ hybridization of quantum dots. By preparing quantum dot coupled DNA (deoxyribonucleic acid) as a probe, according to the traditional fluorescence in situ hybridization method, the hybridization detection of vegetal single copy genes is carried out on interphase nucleuses, chromosomes, DNA fibersor the like. In the method provided by the invention, a direct marking method is adopted, thus an antibody detection process is omitted; and because the quantum dots have high fluorescence intensity and can resist photo-bleaching, the sensitivity and resolution of hybridization are improved, and the vegetal single copy genes can be quickly and effectively located. The method provided by the invention can be widely used for drawing physical maps and analyzing the composition, structure, rearrangement, evolutionary relationship and the like of genomes.
Owner:WUHAN UNIV
Papaya functional centromeric antigen polypeptide and its application
InactiveCN105399805BPromote formationFunction increaseSerum immunoglobulinsImmunoglobulins against plantsNew Zealand white rabbitPhysical Maps
The invention provides a pawpaw functional centromere antigen polypeptide, and applications thereof. The amino acid sequence of pawpaw functional centromere histone CENH3 is represented by SEQ ID No.2; a specific sequence of the N terminal of the amino acid sequence coded based on the pawpaw functional centromere histone CENH3 is used for synthesis of a polypeptide; routine immune-inoculation of New Zealand white rabbits with the polypeptide is carried out, antiserum is prepared, the antibody of CENH3 protein is obtained via antigen affinity purification, and immunofluorescence assay of the antibody is carried out. The antibody of CENH3 protein can be used for cellular localization of pawpaw functional centromere, and can be applied to pawpaw chromatin immunoprecipitation assay sequencing (ChIP-seq), so that the position, size, and sequence information of functional centromeres on pawpaw chromosomes can be determined, obtained physical map information is more complete, and centromere-related information can be provided for pawpaw genome assembly.
Owner:FUJIAN AGRI & FORESTRY UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com