New wheat gene TaMYB3 for regulating synthetization and metabolization of anthocyanin
An anthocyanin and gene technology, applied in genetic engineering, plant genetic improvement, angiosperms/flowering plants, etc., can solve the problems of grape embryogenic callus cells producing anthocyanins and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: Cloning of the TaMYB3 gene
[0031] All experimental materials were purchased from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences. All samples for DNA and RNA extraction were snap-frozen in liquid nitrogen and stored in a -80 freezer after sampling. All nucleotide sequence determinations were performed at Beijing Aoke Biotechnology Co., Ltd. DNA was extracted by CTAB method (Murray and Thompson, 1980), and RNA was extracted by modified Trizon method (Li and Trick, 2005). Plastid extraction was performed using a kit from Beijing Bodatech Bio-Gene Technology Co., Ltd. (China). Nucleotide and protein sequences were aligned using the software Vector NTIAdvance 10.0 (http: / / www.invitrogen.com / ). The protein phylogenetic tree was constructed using the software MEGA 4.0 (http: / / www.megasoftware.net / ). The gene map was drawn using GSDS software (http: / / gsds.cbi.pku.edu.cn / help.php).
[0032] By targeting the MYB-like transcripti...
Embodiment 2
[0033] Example 2: TaMYB3 Bioinformatics Analysis
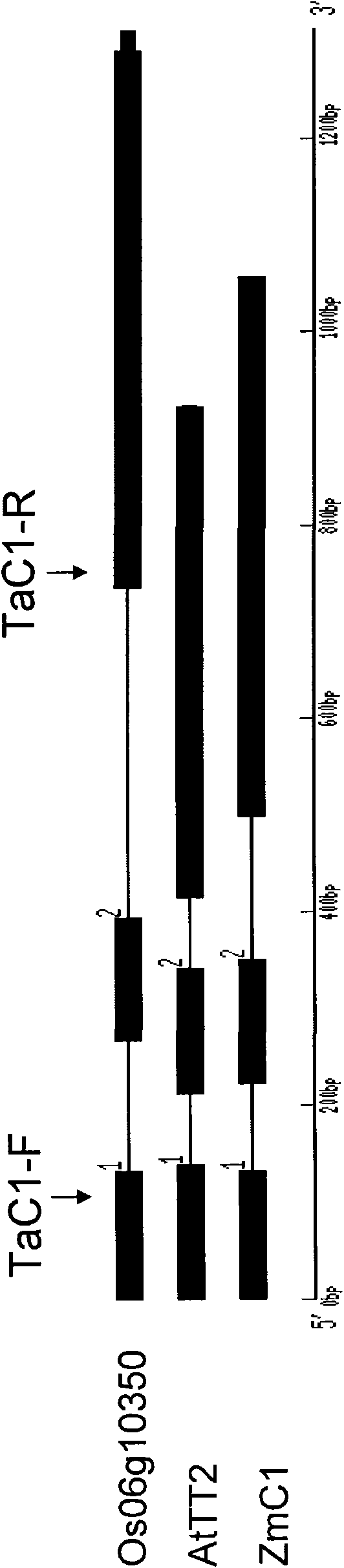
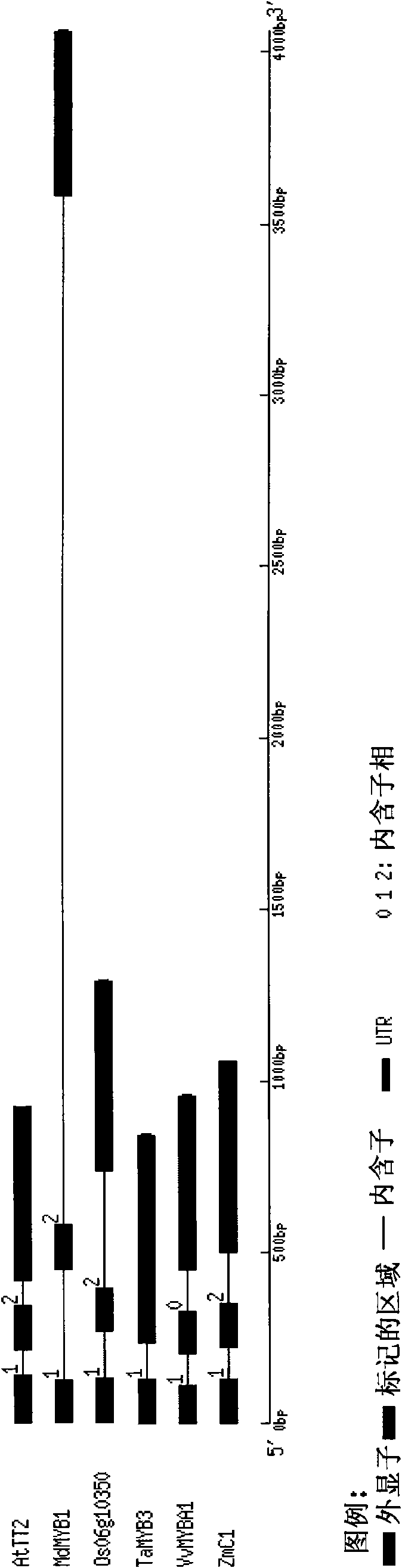
[0034] In order to understand the relationship between TaMYB3 and other similar genes, so as to predict the biological function of TaMYB3, we performed bioinformatics analysis on TaMYB3. Bioinformatics analysis showed that the predicted TaMYB3 protein (SEQ ID NO: 12) had two conserved domains of R2 and R3 with high homology compared with other MYB transcription factors that regulate anthocyanin synthesis ( figure 2 ). The gene structure of TaMYB3 is different from other homologous genes. TaMYB3 has only one intron, while the MYB gene that regulates anthocyanin synthesis and metabolism in maize, rice, Arabidopsis and apple has two introns ( image 3 ). We selected the protein full-length sequences of MYB-like transcription factors known to regulate anthocyanin synthesis and metabolism to construct a phylogenetic tree. In terms of homology, TaMYB3 protein belongs to the MYB class of proteins that regulate anthocyanin synthe...
Embodiment 3
[0035] Example 3: Chromosomal loci of the TaMYB3 gene
[0036]The TaMYB3 chromosomal locus was determined by PCR amplification with TaMYB3 gene-specific primers TaMYB3-F and TaMYB3-R in various cytogenetic materials. TaMYB3 could not be amplified in Urartu wheat IE29-1 (with common wheat A chromosome set, but without B and D chromosome sets) and Junjie wheat As69 (with common wheat D chromosome set, without A and B chromosome sets). TaMYB3 can be stably amplified in materials with B genomes such as Chinese Spring and Langdon. This means that TaMYB3 is not on the A and D chromosome sets, but on the B chromosome set ( Figure 5 .B). TaMYB3 could not be amplified in the genomic DNA of tetraploid wheat Langdon (AABB) which lacked chromosome 4B and added chromosome 4D in Chinese spring. TaMYB3 can be stably amplified in other substitution lines with chromosome 4B, and it is inferred that TaMYB3 is located on chromosome 4B of wheat ( Figure 5 .B). In the Chinese spring 4B chro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com