Method for corresponding peanut genome chromosome sequence map to actual karyotype chromosome number
A chromosome and genome technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of difficulty in using excellent genes in genome research, unclear physical locations, etc., and achieve the effect of promoting research and utilization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
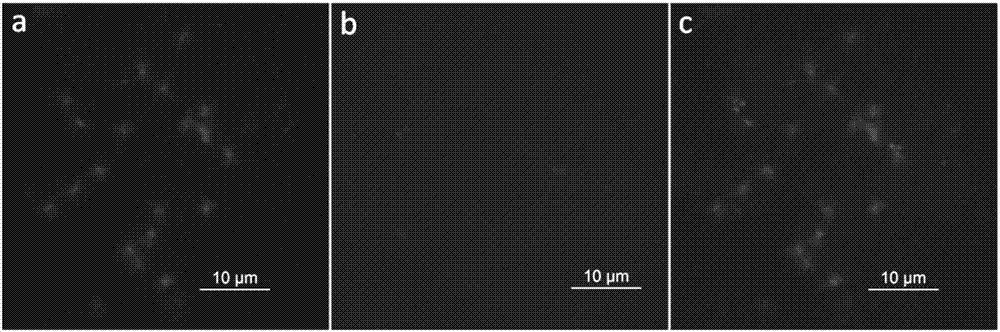
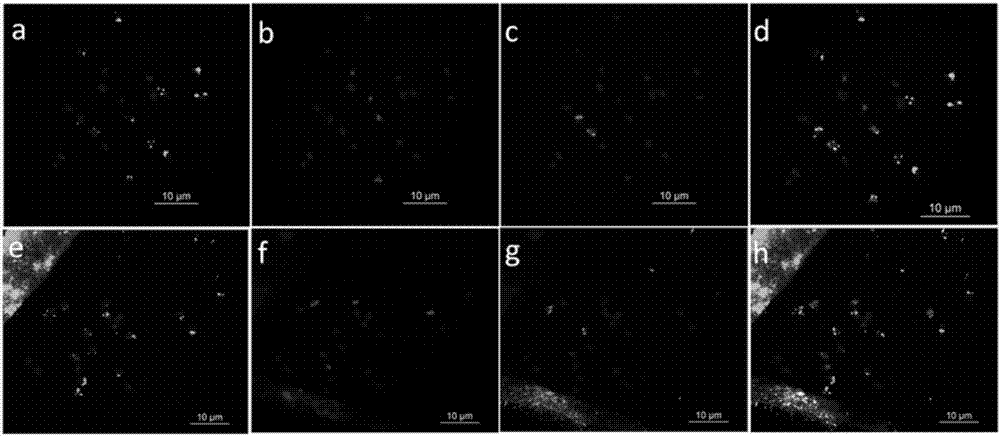
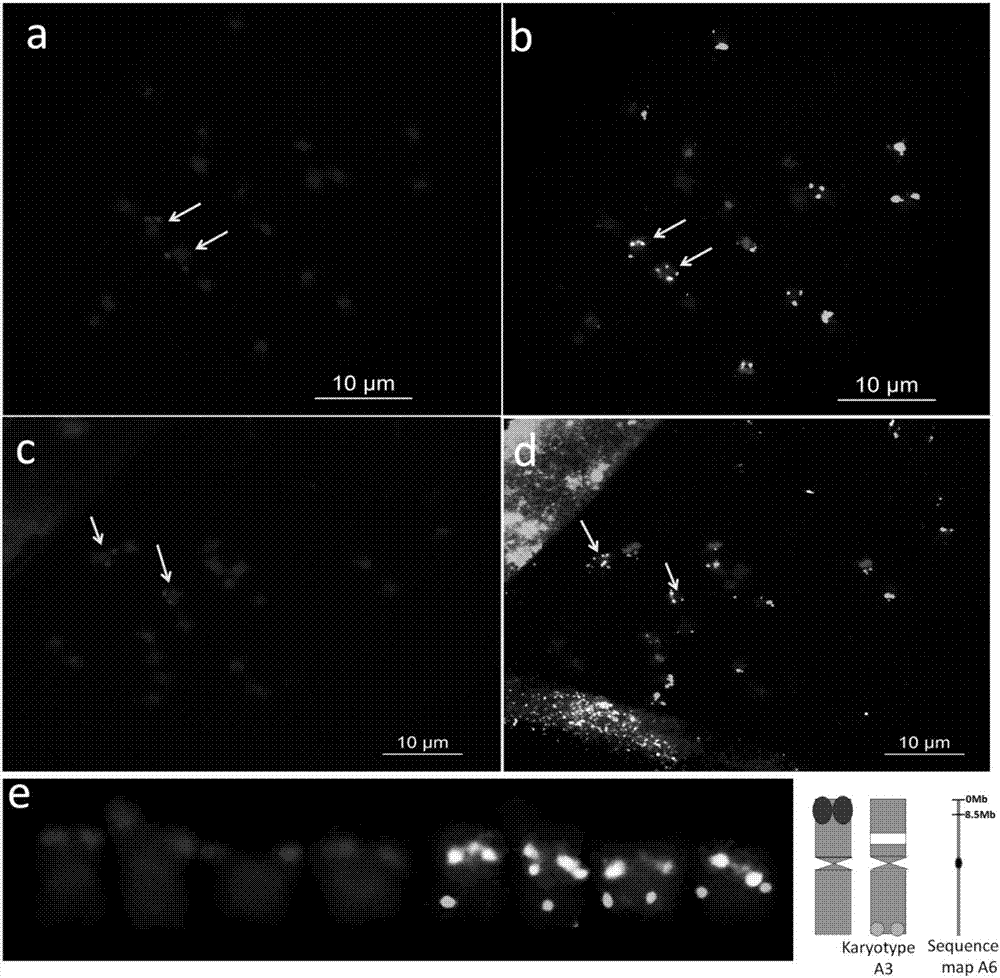
[0053] A method for corresponding the peanut genome chromosome sequence diagram to the actual karyotype chromosome sequence number, comprising the following steps:
[0054] (1) Design of chromosome-specific oligonucleotide (oligo) probes
[0055] Using the draft genome sequence of the peanut wild species A. duranensis published by PeanutBase (https: / / peanutbase.org / gbrowse_aradu1.0), select the 0-8.5 Mb end of the short arm of the gene-enriched region A6 as the target segment. Then design analysis parameters: (a) oligonucleotide length 42-48nt; (b) oligonucleotide density>2oligos / kb; (c) oligonucleotide dTm (DNA melting temperature - hairpin DNA melting temperature)> 10°C; the genome sequence and analysis parameters of the target segment were sent to MYcroarray Company (USA) for bioinformatics analysis, and MYcroarray Company used the RepeatMasker software to analyze the target segment and eliminate the genome repeat sequence of the target segment (http: / / www. repeatmasker.or...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com