Restriction enzyme based whole genome sequencing
A restriction enzyme and restriction technology, applied in the field of sequencing of biological genomes or parts thereof, can solve the problems of fierce competition, heavy, expensive draft genome sequences, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
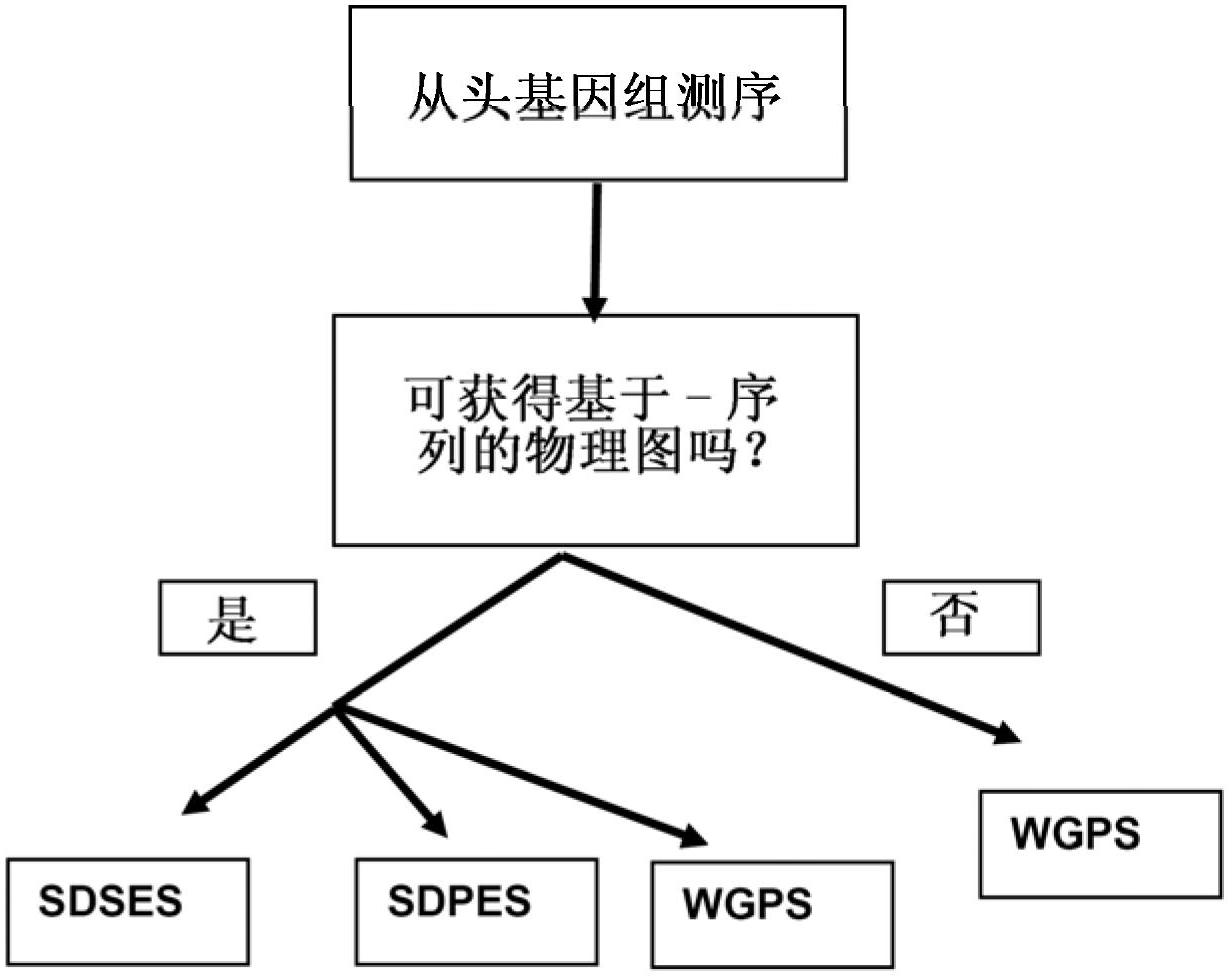
[0195] WGPS embodiment
[0196] The melon BAC library superset 24 is presented together with WGP data from superset 24 to demonstrate successful WGPS on the BAC set by splicing GA paired-end reads linked to enzyme sites.
[0197] 1. Wetlab method
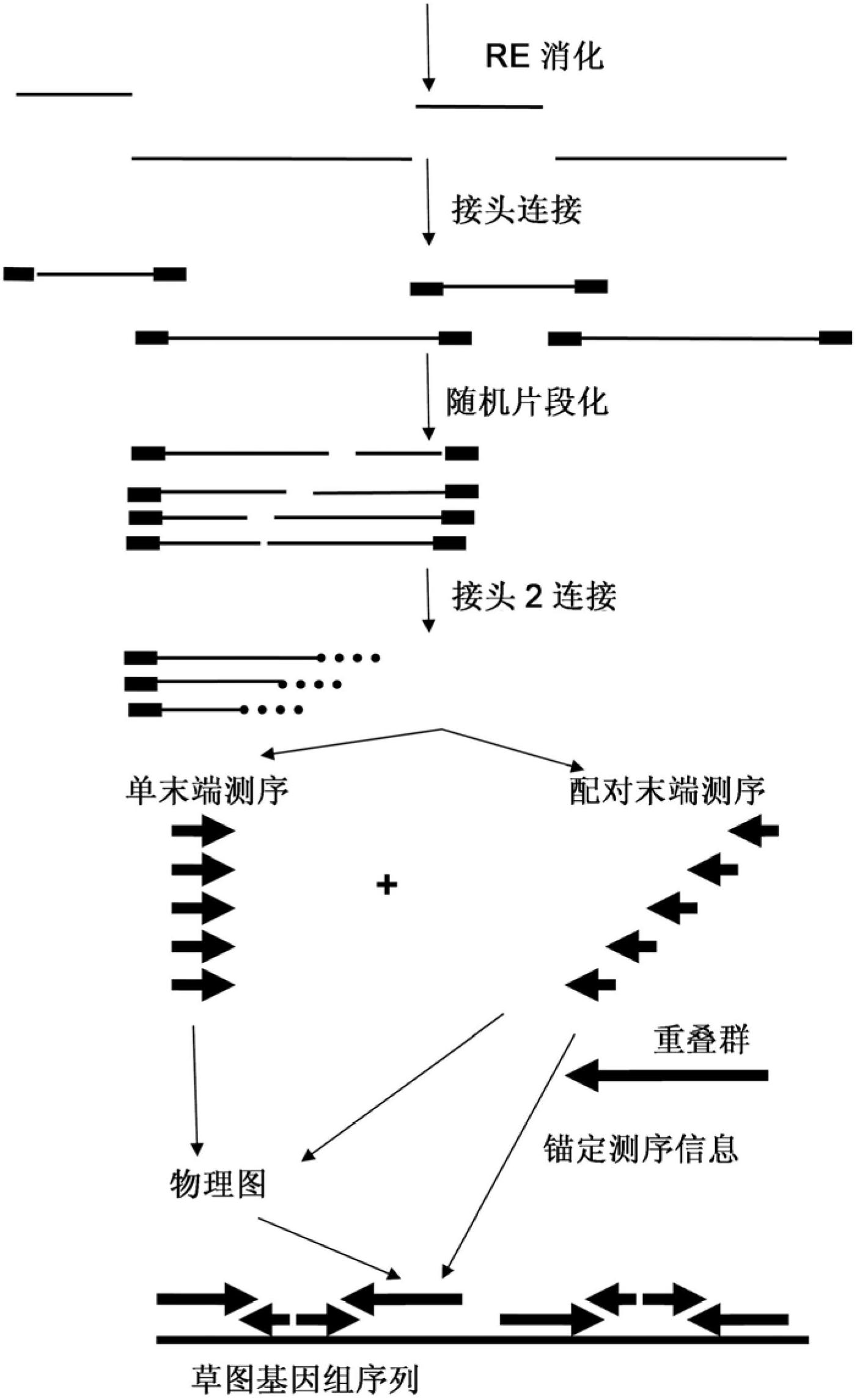
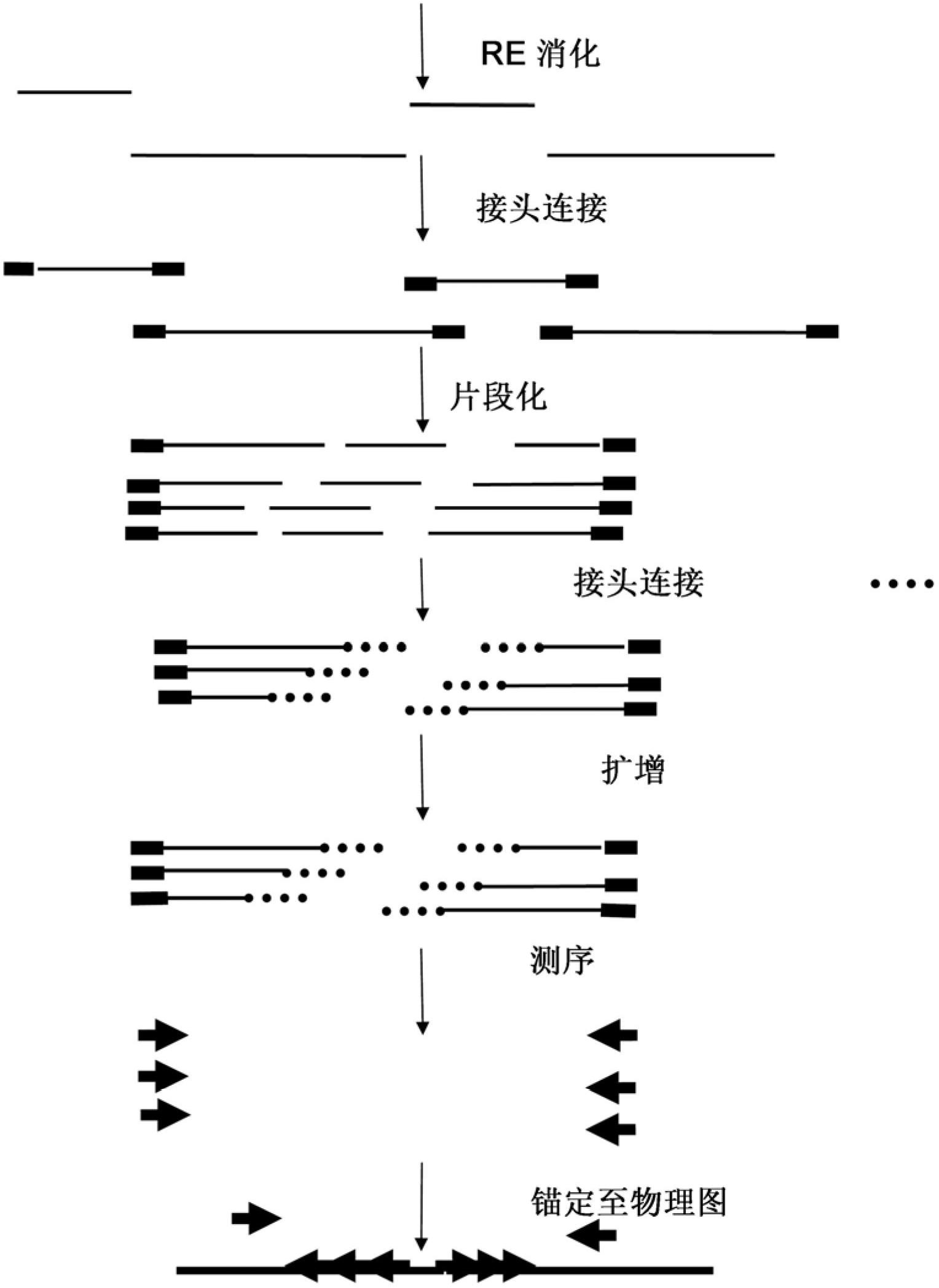
[0198] The method contains the following steps:
[0199] - Digest (individual) BAC pool DNAs using a single enzyme (EcoRI).
[0200] - Ligation of pool-specific EcoRI compatible adapters containing P5 amplification, sequence primer 1 and pool-specific identifier sequences.
[0201] - (optionally pooling RL products from supersets that will be sequenced in a single pass on e.g. an Illumina Genome Analyzer. This is up to a maximum of different set-specific identifiers used in the previous ligation step)
[0202] - Fragmentation of the adapter ligated products into products with a size in the range of 100-1000 bp.
[0203] Fragmented adapter-ligated restriction fragments were blunt-ended and a single A-nt was added to the fragmente...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com