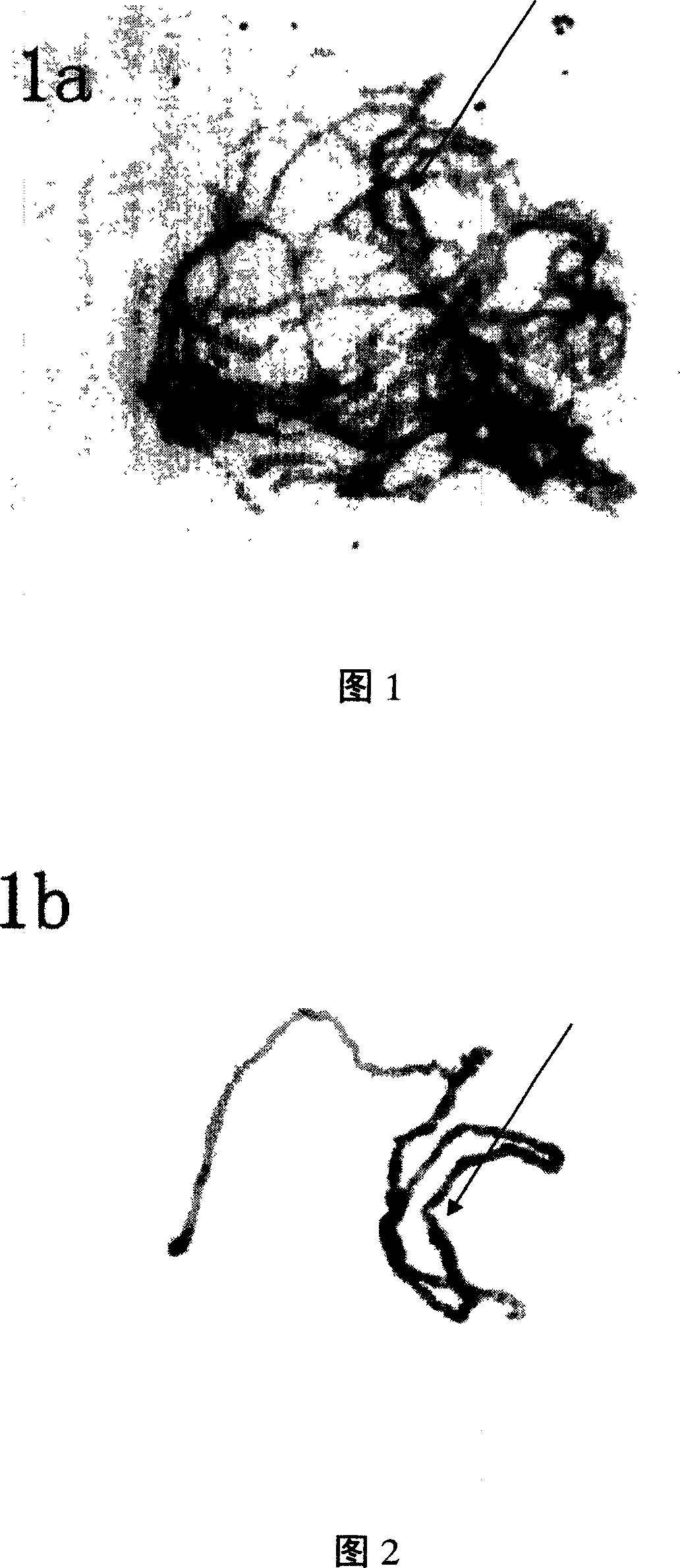
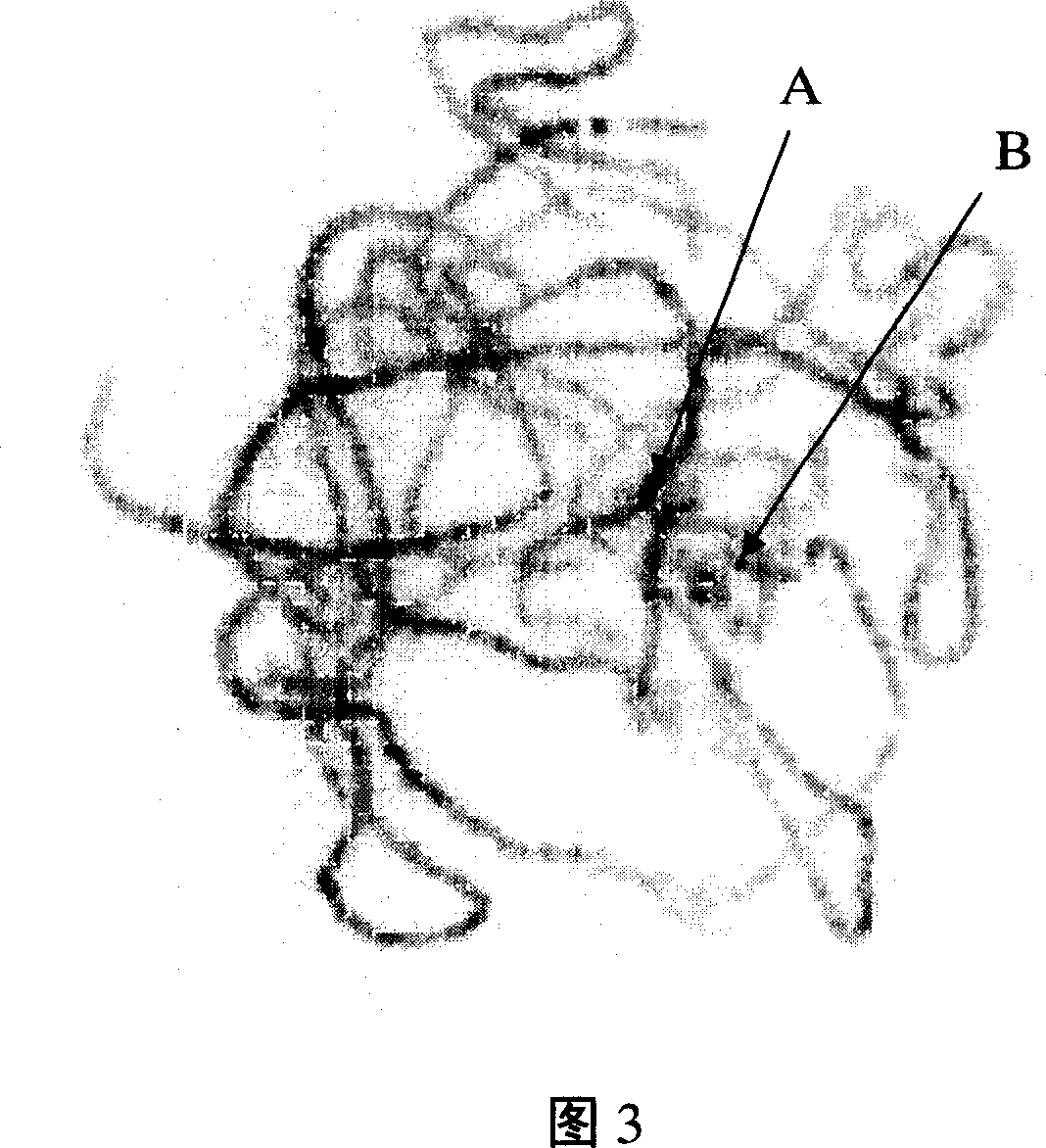
Cotton pachytene chromosome fluorescence in-situ hybridization method
A technique of fluorescence in situ hybridization and hybridization method, which is applied in the field of fluorescence in situ hybridization of cotton pachytene chromosomes, and achieves the effects of improving detection efficiency and resolution.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0029] 2. Probe preparation
[0030] The probe selected by signal multiple amplification fluorescence in situ hybridization is EST sequence G1185, the gene bank number is CA994275, and the primers designed by Oligo6 software are:
[0031] 5'GGATTATGAATGGCGACAAG3' (upstream primer, SEQ ID NO: 1),
[0032] 5'TTTAGAAATCTTCCTCCGTT3' (downstream primer, SEQ ID NO: 2), the length of the PCR product is 934bp;
[0033] The probes selected by two-color fluorescence in situ hybridization were 18S rDNA and 5S rDNA genes, and the primers were designed using Oligo6 software. The primers designed for the 18S rDNA gene were:
[0034] 5'TGTGAAACTGCGAATGGCTC3' (upstream primer, SEQ ID NO: 3),
[0035] 5'ACAAAGGGCAGGGACGTAGT3' (downstream primer, SEQ ID NO: 4), the length of the PCR product is 1500bp;
[0036] The primers designed for the 5S rDNA gene are:
[0037] 5'GAGAGTAGTACAACGATGGG3' (upstream primer, SEQ ID NO: 5),
[0038] 5'GGAGTTCTGATGGGATCCGG3' (downstream primer, SEQ ID NO: 6),...
example 1
[0044] Example 1 Signal Multiplex Amplified Fluorescence In Situ Hybridization
[0045]Before hybridization, dry the prepared sheet in an oven at 60°C overnight; add a few drops of 100 μg / μL DNase-free RNase, cover the sheet, and treat it at 37°C for 1 hour, and use 2×SSC (0.3mol / L NaCl and 0.03mol / L trisodium citrate) buffer solution, rinse the coverslip with 2×SSC buffer solution, and wash the preparation piece with 2×SSC buffer solution for 3×5 minutes (washing 3 times, 5 minutes each time); drop Add a few drops of 0.01% (suitable for somatic cells) or 1% (suitable for pollen mother cells) Pepsin (prepared in 10mM HCl), cover the cover slip, incubate at 37°C for 1 hour, rinse the cover slip with 2×SSC buffer, 2× SSC buffer was washed for 2×5 minutes; 2% PVP40 and 4g / L of active carbon were treated for 1 hour, and 2×SSC buffer was washed for 3×5 minutes; 4% paraformaldehyde (50mM MgCl 2 1 × PBS preparation) fixed at 37°C for 10 minutes; washed with 2 × SSC buffer for 3 × 5...
example 2
[0049] Example 2 Two-color fluorescence in situ hybridization
[0050] Before hybridization, dry the prepared slices in an oven at 60°C overnight; add a few drops of 100 μg / μL DNase-free RNase (prepared in 2×SSC buffer), cover with a cover slip, and treat at 37°C for 1 hour, 2×SSC buffer Rinse off the cover slip, wash with 2×SSC for 3×5 minutes; add a few drops of 1% Pepsin (prepared in 10mM HCl), cover the cover slip, incubate at 37°C for 1 hour, rinse the cover slip with 2×SSC buffer, 2 × SSC buffer wash 2 × 5 minutes; transfer to 4% paraformaldehyde (50mM Mgcl 2 1 × PBS preparation) fixed at 37 ° C for 10 minutes; 2 × SSC buffer wash 3 × 5 minutes; quickly transferred to 70%, 90%, 100% ethanol gradient dehydration step by step for 3 minutes, air-dried slides.
[0051] The hybridization mixture must be newly prepared before use, and its composition is as follows: 50% deionized formamide (used to prevent damage and shedding of chromosome shape and structure caused by high t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com