Whole genome sequencing data calculation interpretation method
A whole-genome sequencing and data computing technology, which is applied in computing, electrical digital data processing, special data processing applications, etc., can solve the problems of limited GPU hardware structure characteristics, high energy consumption, large area, etc., to achieve rich diversity, Speed up processing and improve real-time effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
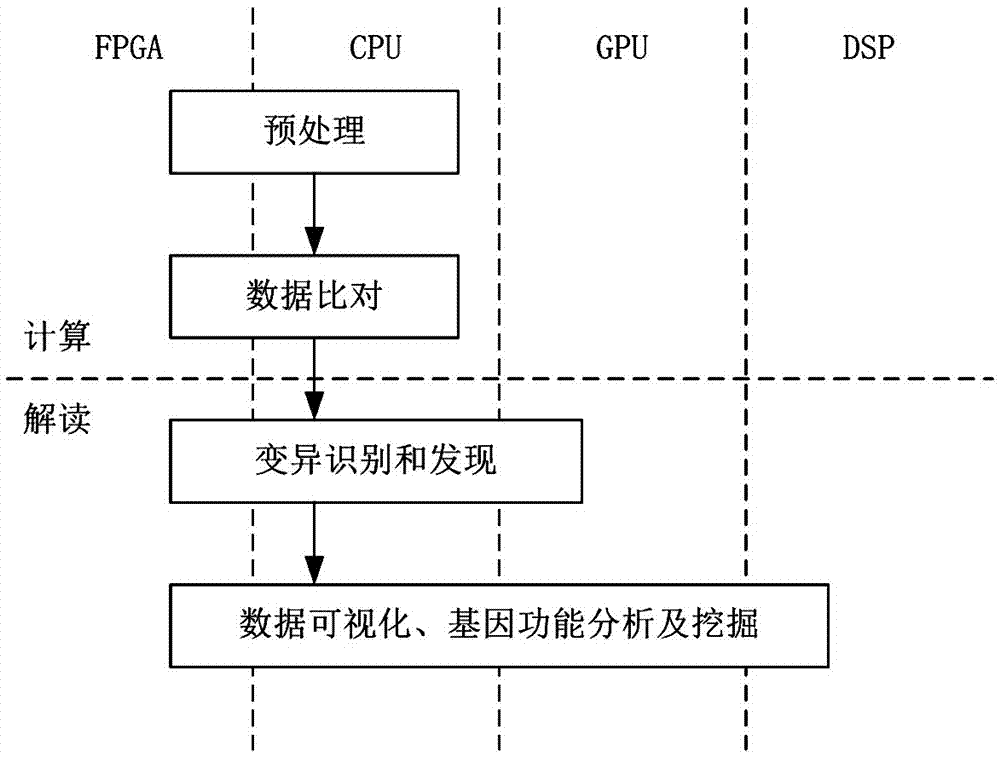
[0054] like figure 1 As shown, the implementation steps of the calculation and interpretation method for whole genome sequencing data in this embodiment include:
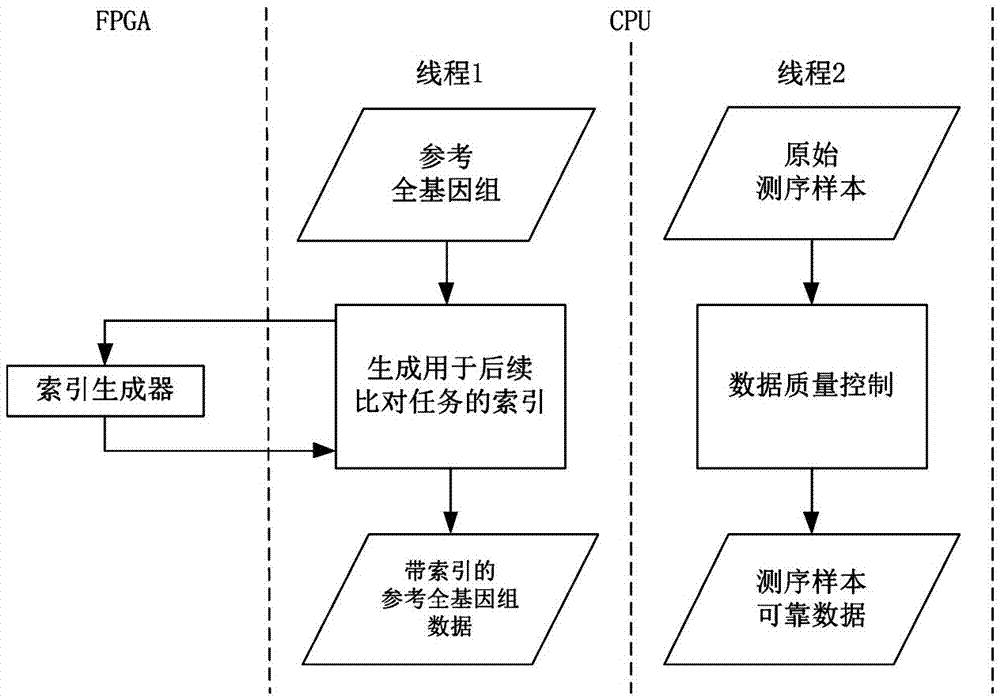
[0055] 1) Input the reference whole genome data and original sequencing sample data for whole genome sequencing; preprocess the original sequencing sample data based on CPU to obtain reliable data of sequencing samples; The genome data is preprocessed to obtain the reference whole genome data with index as the comparison object, and the CPU calls the index generator implemented by FPGA hardware to accelerate the generated index; if the reference whole genome data does not need preprocessing, directly The original indexed reference whole genome data is used as the comparison object; this step requires the use of two processors, CPU and FPGA;
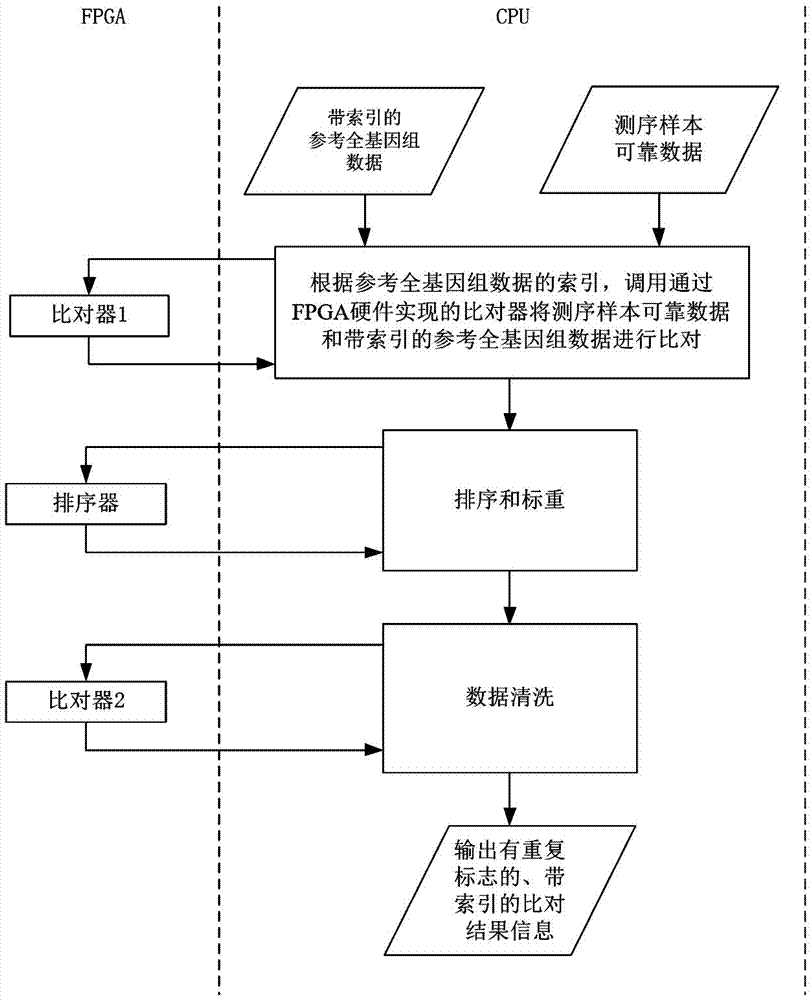
[0056] 2) The CPU calls the FPGA to accelerate the alignment between the reliable data of the sequencing sample and the reference whole genome data with an index (alignment), ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com