Detection method of vegetal single copy genes
A single-copy gene and detection method technology, applied in biochemical equipment and methods, microbial measurement/inspection, material excitation analysis, etc., can solve problems such as short DNA fragments, single-copy detection, difficulty of quantum dots entering the nucleus, etc. , to achieve high fluorescence intensity, improve sensitivity and resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
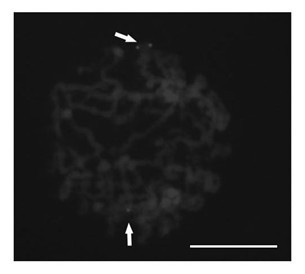
[0024] 1) Prepare maize interphase nuclei and metaphase chromosomes on glass slides;
[0025] 2) Synthesis of Octylamine-modified polyacrylic acid coated CdSe / ZnS quantum dots (Octylamine-modified polyacrylic acid coated CdSe / ZnS quantum dots, OPA-coated CdSe / ZnS QDs);
[0026] 3) According to the corn fatty aldehyde dehydrogenase gene, design and synthesize a pair of oligonucleotide primers, and connect the 5' end of one of the primers to 5'-NH 2 -(CH 2 ) 6 -TTTTTT, and through amino (-NH 2 ) and the carboxyl group (-COOH) on the quantum dots react chemically and couple to the surface of the quantum dots;
[0027] 4) Add the oligonucleotide primer coupled with quantum dots together with another primer and corn genomic DNA into the polymerase chain reaction system, and obtain a 480bp DNA molecule coupled with quantum dots by polymerase chain reaction ;
[0028] 5) Separate the product obtained by polymerase chain reaction by agarose gel electrophoresis to obtain a single ...
Embodiment 2
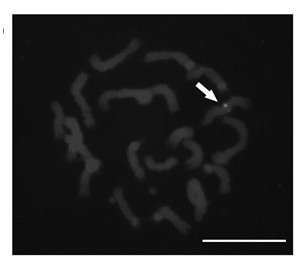
[0034] 1) Prepare maize chromatin fibers on a glass slide;
[0035] 2) Synthesis of Octylamine-modified polyacrylic acid coated CdSe / ZnS quantum dots (Octylamine-modified polyacrylic acid coated CdSe / ZnS quantum dots, OPA-coated CdSe / ZnS QDs);
[0036] 3) Design and synthesize a pair of oligonucleotide primers based on the receptor-like protein kinase 2 gene related to maize somatic embryogenesis, and connect the 5' end of one of the primers to 5'-NH 2 -(CH 2 ) 6 -TTTTTT, and through amino (-NH 2 ) and the carboxyl group (-COOH) on the quantum dots react chemically and couple to the surface of the quantum dots;
[0037] 4) Add the quantum dot-coupled oligonucleotide primer together with another primer and corn genomic DNA into the polymerase chain reaction system, and obtain the quantum dot-coupled 520bp DNA molecule through polymerase chain reaction ;
[0038] 5) Separate the product obtained by polymerase chain reaction by agarose gel electrophoresis to obtain a single ...
Embodiment 3
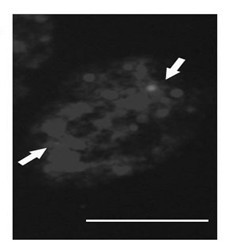
[0044] 1) Prepare parsley metaphase chromosomes on glass slides;
[0045] 2) Synthesis of Octylamine-modified polyacrylic acid coated CdSe / ZnS quantum dots (Octylamine-modified polyacrylic acid coated CdSe / ZnS quantum dots, OPA-coated CdSe / ZnS QDs);
[0046] 3) According to the chalcone synthase gene of parsley, design and synthesize a pair of oligonucleotide primers, and connect the 5' end of one of the primers to 5'-NH 2 -(CH 2 ) 6 -TTTTTT, and through amino (-NH 2 ) and the carboxyl group (-COOH) on the quantum dots react chemically and couple to the surface of the quantum dots;
[0047] 4) Add the oligonucleotide primer coupled with quantum dots together with another primer and parsley genomic DNA into the polymerase chain reaction system, and obtain the 490bp DNA coupled with quantum dots by polymerase chain reaction molecular;
[0048] 5) Separate the product obtained by polymerase chain reaction by agarose gel electrophoresis to obtain a single 490bp DNA molecule c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com