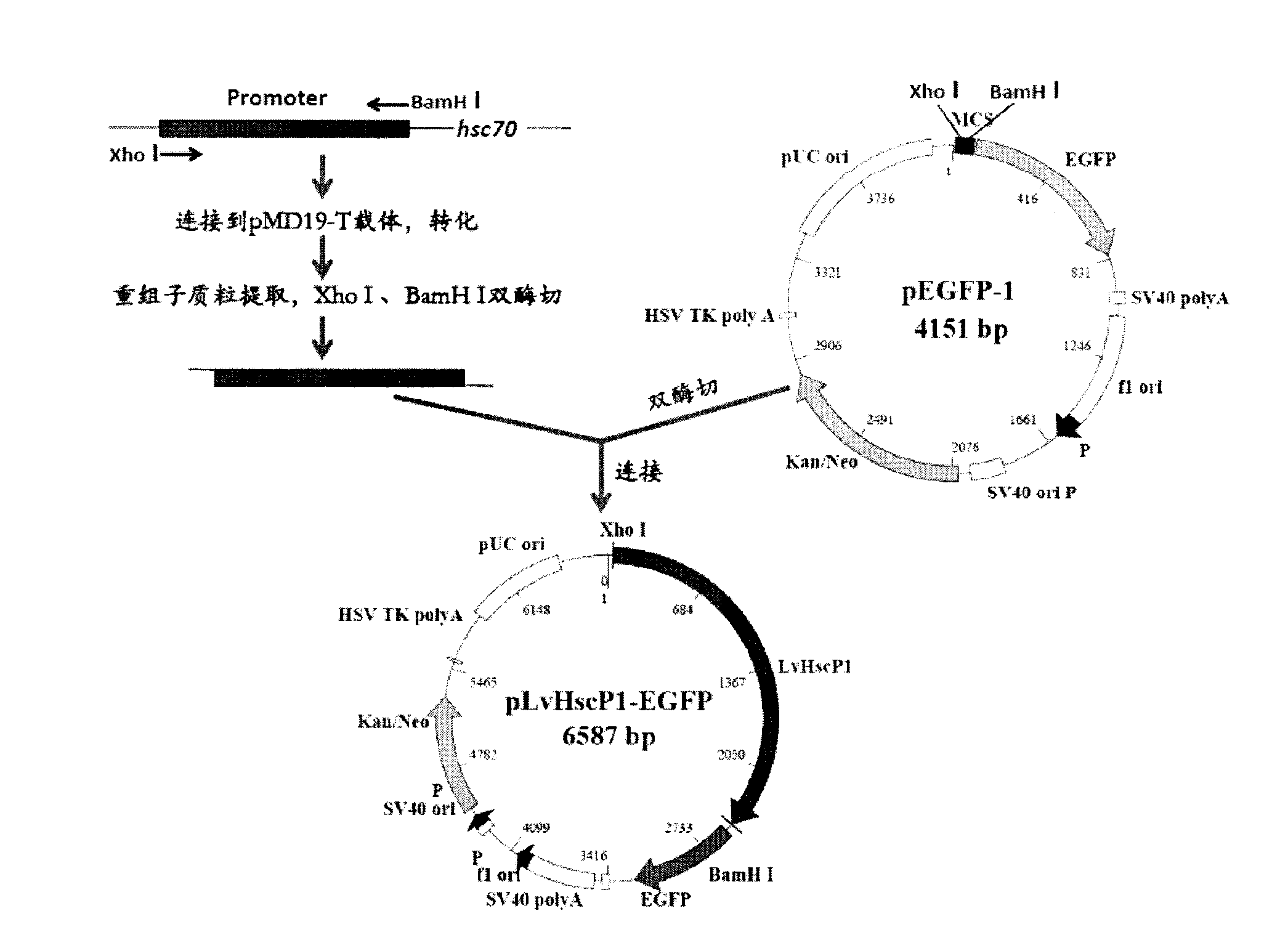
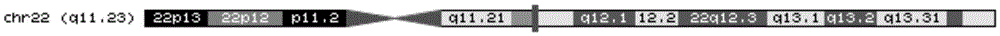Patents
Literature
60 results about "Bac clone" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The BAC clones are added to bacterial cells, usually E. coli. The bacteria are then spread on nutrient rich plates that allow only the bacteria that carry BAC clones to grow. The bacteria grow rapidly, resulting in lots of bacterial cells, each containing a copy of the BAC clone. After they have grown,...
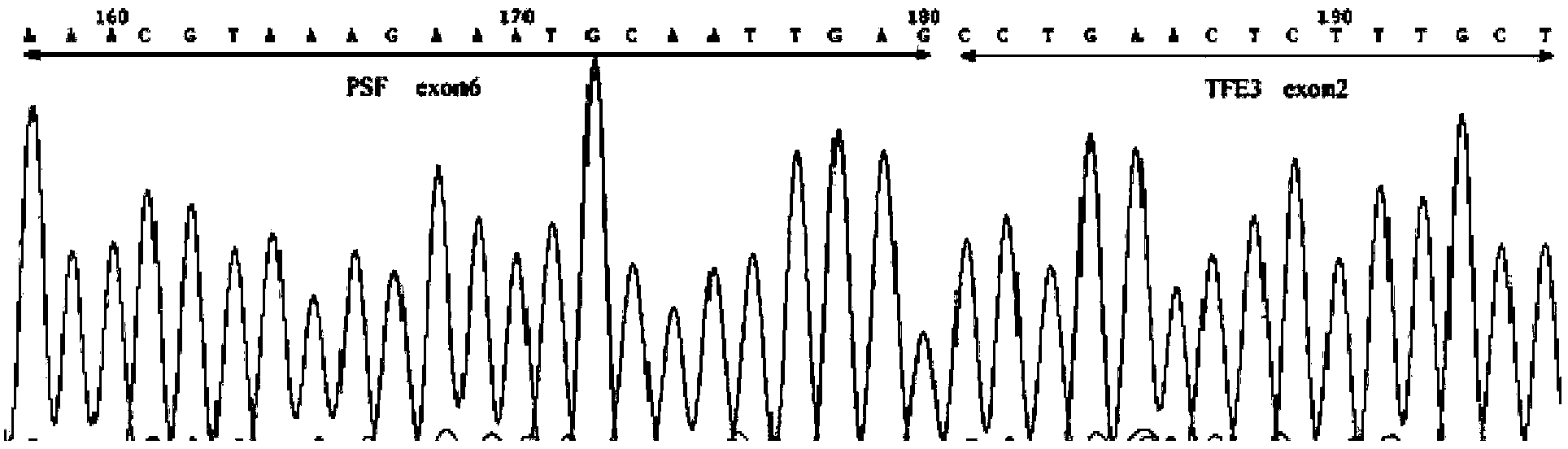
Probe combination for diagnosing Xp11.2 translocation renal cancer and application thereof
InactiveCN102424848AImprove accuracyIncrease success rateMicrobiological testing/measurementDNA/RNA fragmentationBac cloneFluorescence
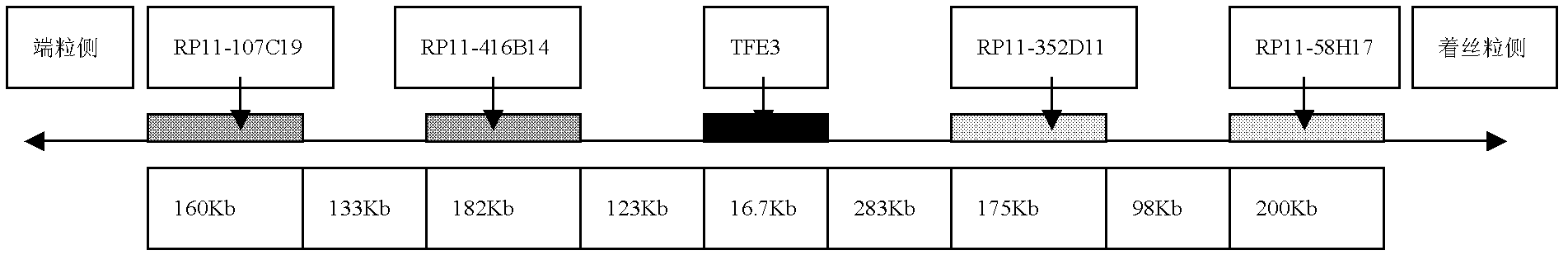
The invention discloses a probe combination for diagnosing Xp11.2 translocation renal cancer and an application thereof, which belong to the field of fluorescent in situ hybridization probe application. The probe combination is BAC cloning fragments RP11-58H17, RP11-352D11, RP11-416B14 and RP11-107C19. The specificity and the sensibility of the Xp11.2 translocation renal cancer diagnosis by the probe combination respectively reach 100 percent, convenience, high speed and reliability are realized, in addition, the success ratio is high, and the probe combination can be used for preparing Xp11.2 translocation renal cancer diagnosis reagents.
Owner:NANJING GENERAL HOSPITAL NANJING MILLITARY COMMAND P L A
Probe set for diagnosing Xp11.2 ectopic perivascular epithelioid cell neoplasm and application of probe set
ActiveCN104212889AImprove accuracyIncrease success rateMicrobiological testing/measurementDNA/RNA fragmentationBac cloneFluorescence
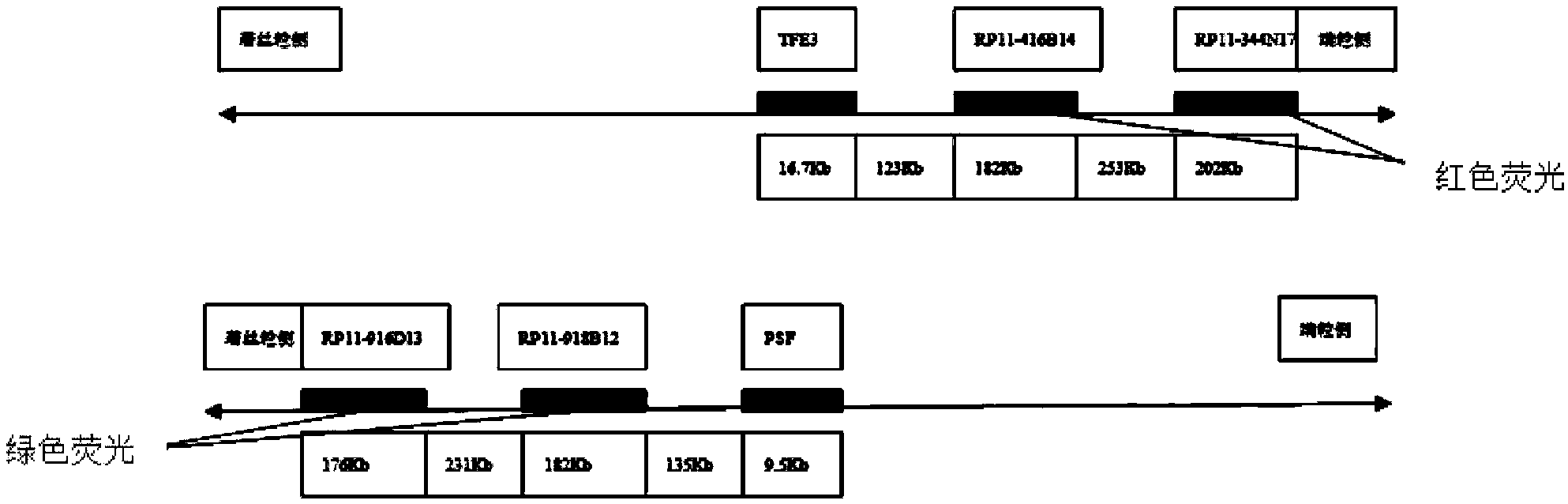
The invention belongs to the application field of fluorescence in situ hybridization probes and discloses a probe set for diagnosing Xp11.2 ectopic perivascular epithelioid cell neoplasm and application of the probe set. The probe set includes BAC clonal fragments RP11-918B12, RP11-916D13, RP11-416B14 and RP11-344N17. The probe set is capable of diagnosing the specificity and the sensibility of Xp11.2 ectopic PEComa 100%, is convenient, quick and reliable, and can ensure a high success rate. And moreover, the probe set can be used for preparing an Xp11.2 ectopic PEComa diagnostic kit.
Owner:NANJING GENERAL HOSPITAL NANJING MILLITARY COMMAND P L A
Reagent for detecting copy number of EGFR gene and ploidy of chromosome 7
ActiveCN101899504AHelps with sensitivityAids in prognostic judgmentMicrobiological testing/measurementFluorescence/phosphorescenceTyrosine-kinase inhibitorPloidy
The invention discloses a reagent for detecting a copy number of an EGFR gene and the ploidy of a chromosome 7. The reagent comprises a fluorescence probe RP11-339F13 used for detecting the copy number of the EGFR gene and a fluorescence probe RP11-144H20 used for detecting the ploidy of the chromosome 7, wherein the RP11-339F13 is cloned by BAC, which covers the whole EGFR gene, and marked with rhodamine fluorescein; and RP11-144H20 is cloned by specific BAC of the centromere of the chromosome 7 and marked with fluorescein isothiocyanate. The reagent can quickly, simply and conveniently detect the copy number of the EGFR gene and the ploidy of the chromosome 7, contributes to the judgment of medicament susceptibility and prognostic judgment, and has great significance for the guidance of use of molecular targeted medicament tyrosine kinase inhibitors (TKIs) of tumors, such as non-small cell lung cancer and the like.
Owner:合肥艾迪康医学检验实验室有限公司
Method for cloning rice auxin induced protein gene
InactiveCN101492671AHigh homologySimple implementation stepsFermentationPlant genotype modificationMutantDNA Intercalation
The invention relates to a method for using rice auxin to induce protein genes to be cloned, which is characterized in that the implementation steps comprise: (1) the preparation of a rice transformation receptor; (2) the genetic transformation of the rice; (3) the screening of kanamycin-resistant callus tissue and the regeneration of plants; (4) the screening of the mutant of a T-DNA inserted progenies; (5) Tail-PCR; (6) the comparison and analysis of sequences on the Internet. In the invention, a rice mutant with a short plant height is obtained when carrying out rice functional genome research by using T-DNA label method; the lateral neighboring sequences of the mutant are researched by using TAIL-PCR technology; meanwhile, the position where the mutant T-DNA inserts the rice genome is arranged on the No. 4 chromosome of the rice by the comparison on the databases of NCBI and TIGR on the Internet; moreover, the rice BAC clone (OSJNBa0084K01) of the position is found out. The T-DNA is inserted between the two genes of the clone by analyzing the clone. Known functional genes with a very high homology with BAC cloning code amino acid sequence are forecasted by the sequence comparison on the Internet.
Owner:TIANJIN AGRICULTURE COLLEGE
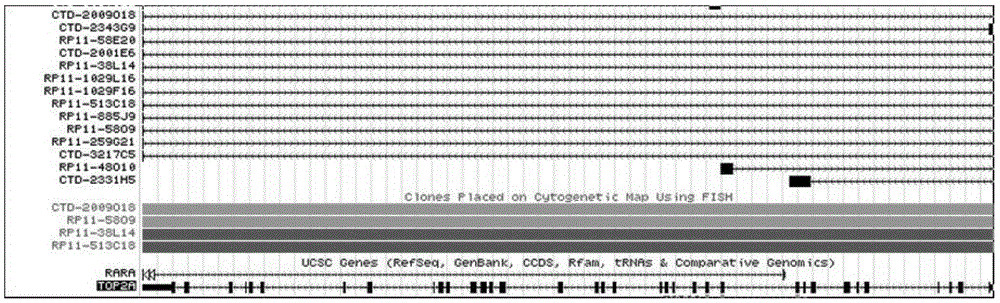
PML gene and RARA gene detection probe, preparation method thereof and reagent kit
InactiveCN105420397AGood repeatabilityEasy to filterMicrobiological testing/measurementDNA/RNA fragmentationBac clonePlasmid dna
The invention relates a PML gene and RARA gene detection probe and a preparation method thereof. The method comprises the following steps that at least one of RP11-832J18, CTD-2529B11, RP11-756N20 and RP11-1031J4 and a selected BAC cline for an RARA gene is at least one of CTD-2360L10, RP11-737D6, CTD-3087O22, RP11-48O10 and CTD-2134K5 are included; a plasmid DNA is obtained; labeling is performed. The invention further discloses a reagent kit containing the PML gene and RARA gene detection probe for detecting acute promyelocytic leukemia PML and RARA fusion genes. The optimal PML gene and RARA gene detection probe is obtained through screening, the signal counting row is accurate and rapid, and the result repeatability is good.
Owner:GUANGZHOU LBP MEDICINE SCI & TECH
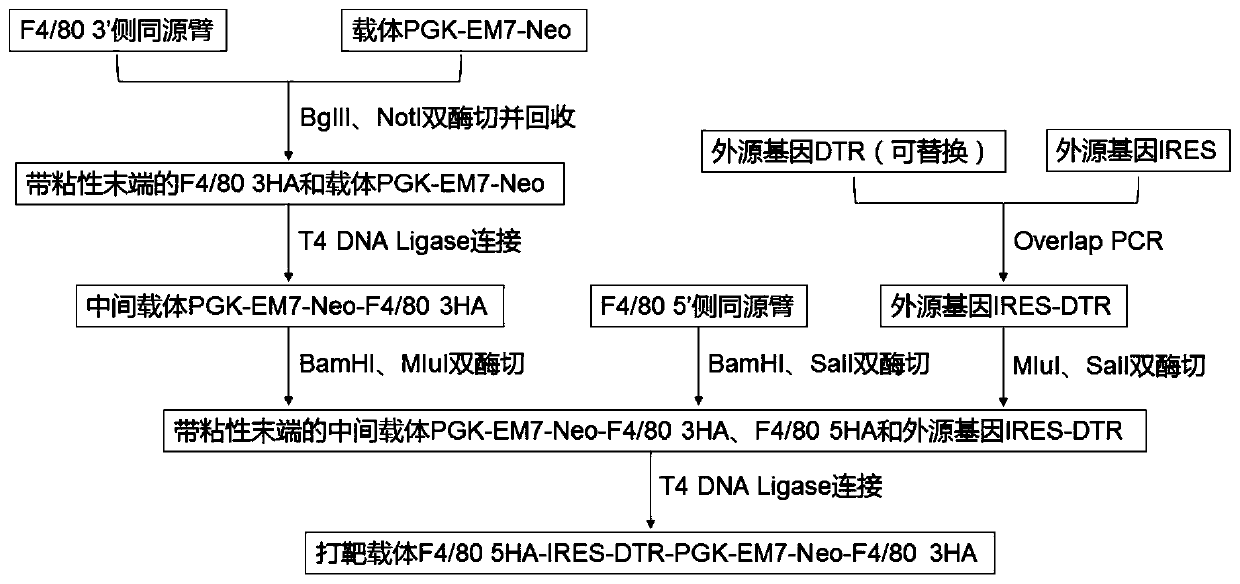
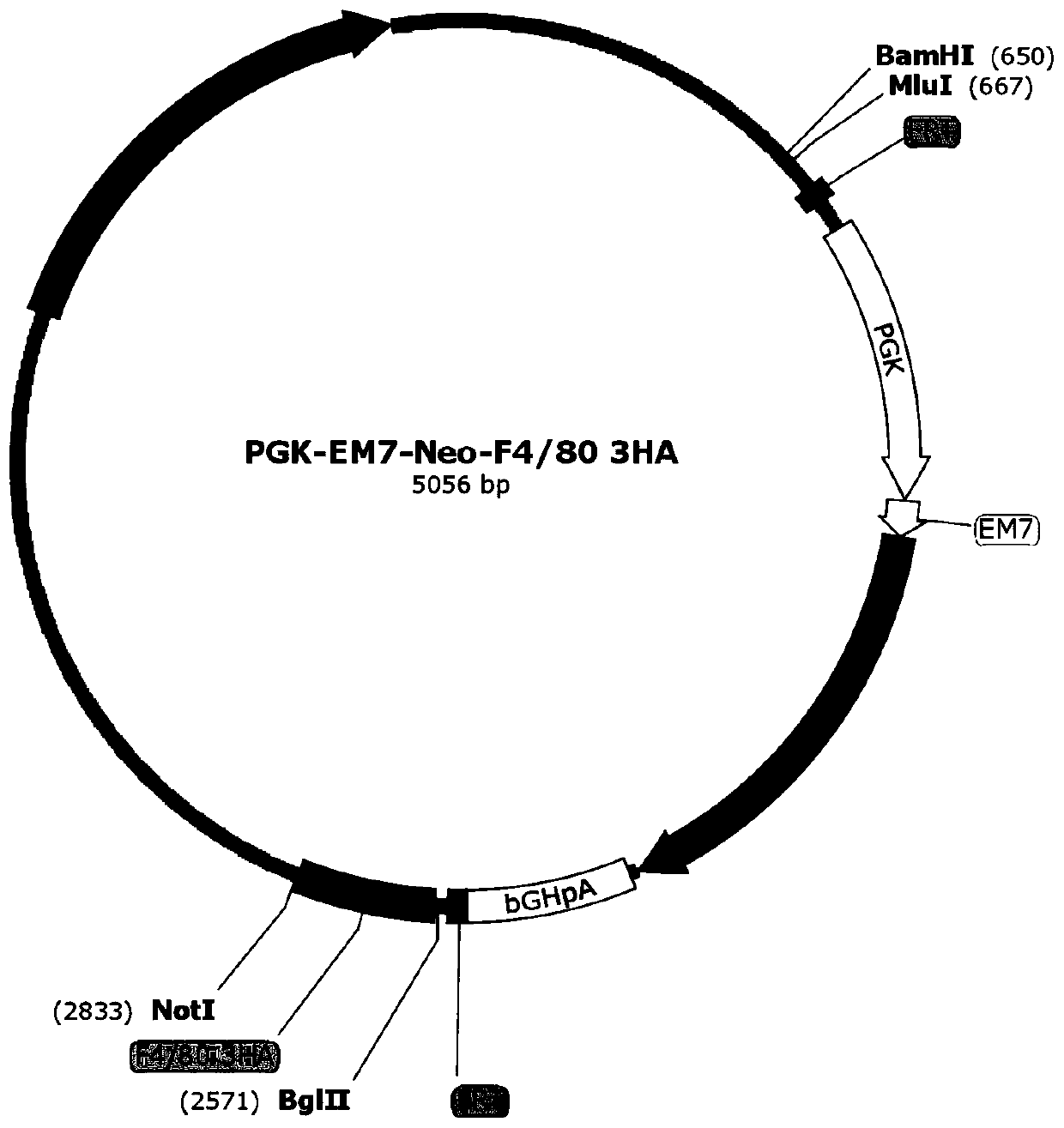
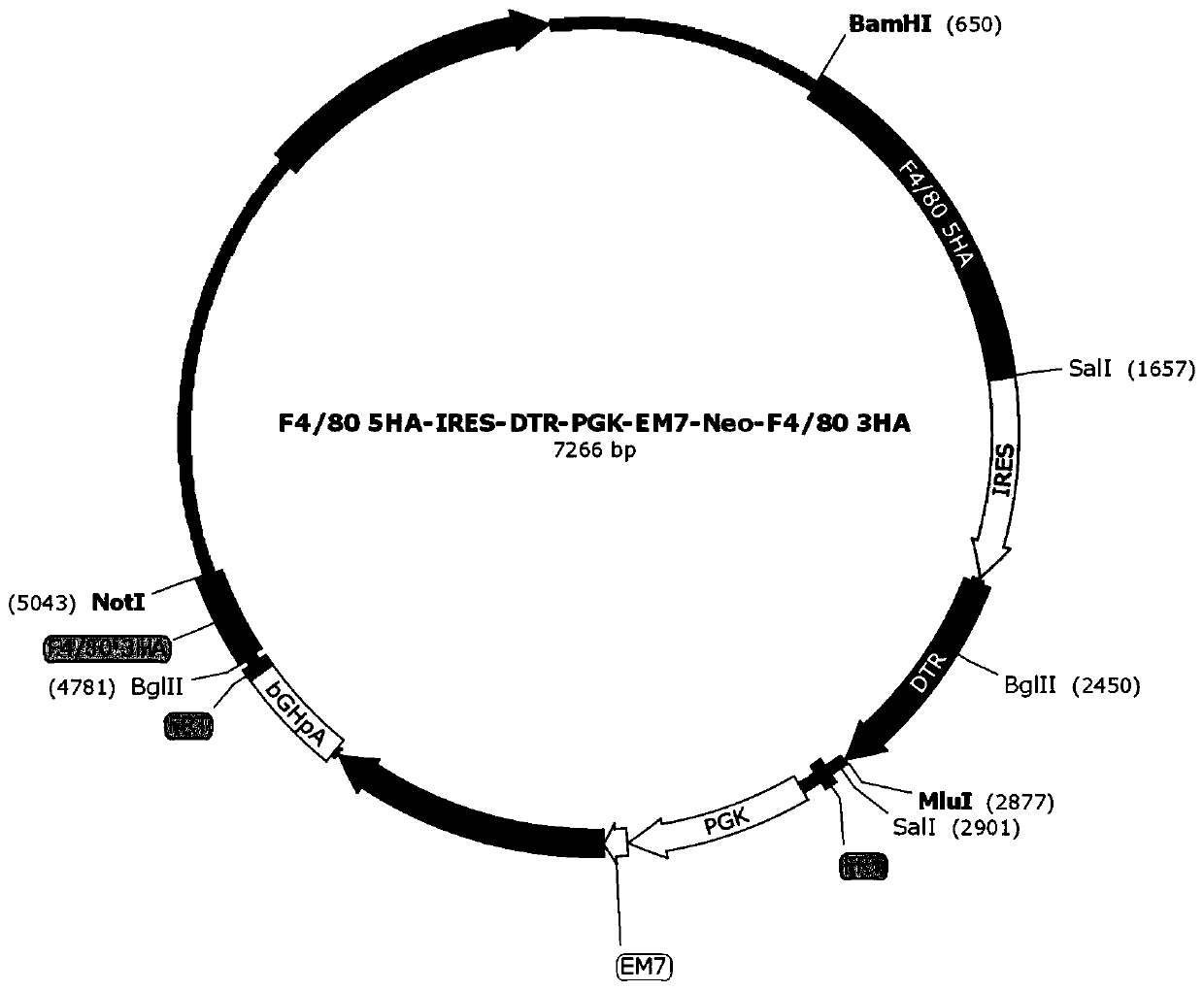
Targeted targeting vector, and method and application for targetedly integrating foreign gene to 22nd position of mouse F4/80 exon to construct BAC clone
PendingCN110938651AEfficient integrationDoes not affect functionStable introduction of DNANucleic acid vectorBac cloneEmbryo
The present invention belongs to the technical field of biology and discloses a targeted targeting vector, and a method and an application for targetedly integrating a foreign gene to a 22nd positionof a mouse F4 / 80 exon to construct BAC clone. A sequence of the targeted targeting vector is shown as SEQ ID NO.14 and named as a vector F4 / 80 5HA-IRES-DTR-PGK-EM7-Neo-F4 / 80 3HA. The present inventionprovides a new site sequence capable of achieving targeted integrating, the site can be used to construct the targeting vector for inserting the foreign gene, the targeting vector is verified to be capable of efficiently integrating the foreign gene into the mouse F4 / 80 BAC clone F4 / 80 exon 22th site, and the subsequently obtained targetedly inserted foreign gene F4 / 80 BAC can be used to construct mouse embryonic stem cells with the targetedly inserted foreign gene, thereby establishing a foundation for constructing a transgenic cell line and mice.
Owner:THE FIRST AFFILIATED HOSPITAL ZHEJIANG UNIV COLLEGE OF MEDICINE
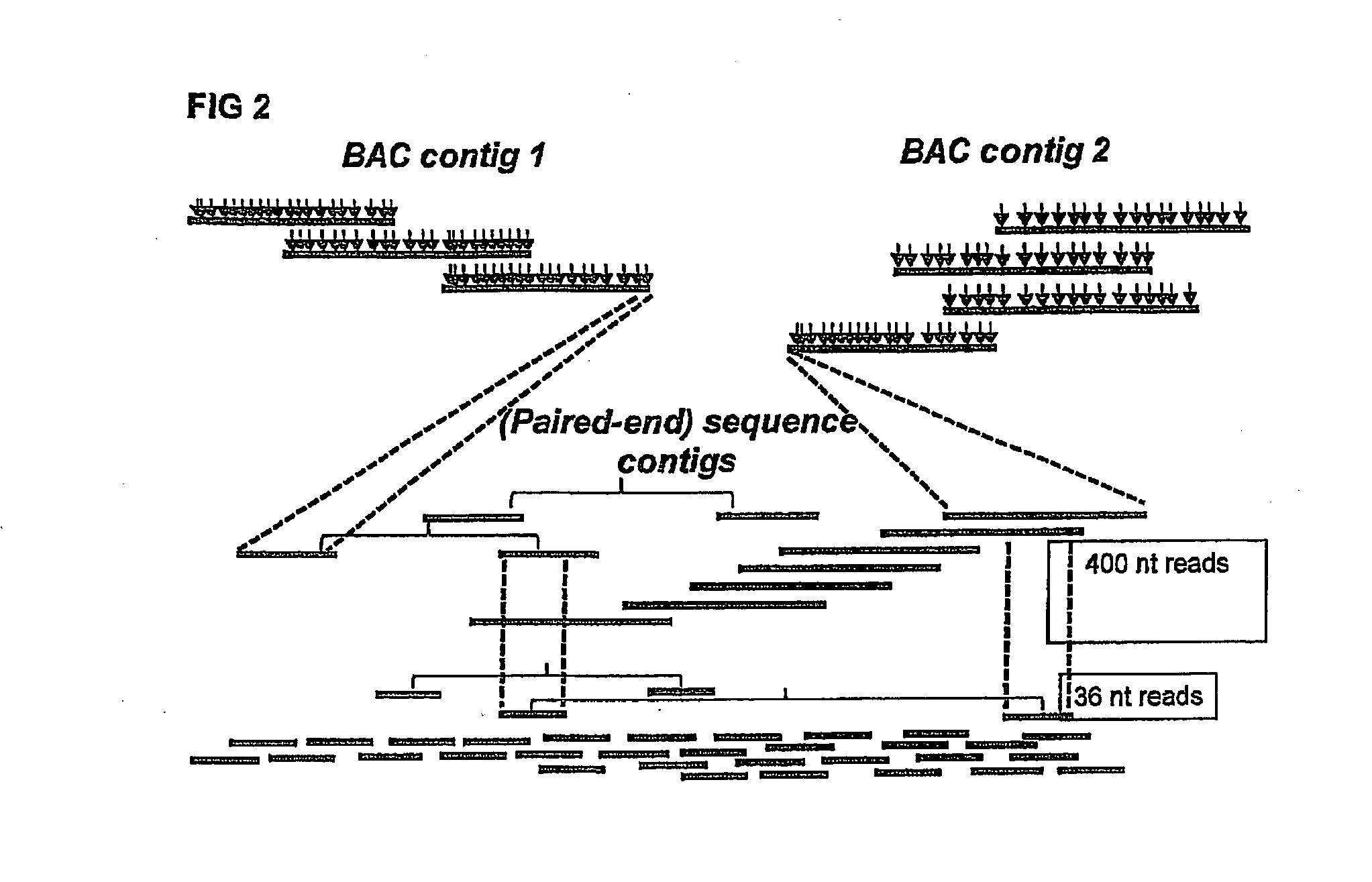
Novel genome sequencing strategies
InactiveUS20130196859A1Increased length and densityQuality improvementMicrobiological testing/measurementLibrary member identificationBac cloneGenomic sequencing
The invention relates to a method for the determination of a genome sequence comprising the steps of providing a physical map of a sample genome by sequencing fragment ends of pooled BAC clones; providing a set of sequence reads from a sample genome generating a contig of the physical map and the sequence reads.
Owner:KEYGENE NV
HER-2 gene and / or TOP2A gene detection probe and preparation method and kit
InactiveCN105524990AStrong specificityHigh sensitivityMicrobiological testing/measurementDNA preparationBac cloneTOP2A Gene
The present invention relates to a HER-2 gene and / or TOP2A gene detection probe and a preparation method thereof, and the method comprises the following steps: selecting BAC aiming at HER-2 gene to clone into RP11-1021P11, RP11-62N23 and RP11-1044P23, or selecting BAC to clone into RP11-98J2, RP11-1065L22 and CTD-2327I15; and / or selecting BAC aiming at TOP2A gene to clone into RP11-1152A10 and CTD-3217C5, or selecting BAC to clone into RP11-737D6, CTD-3087O22, RP11-48O10 and CTD-2134K5; obtaining plasmid DNA; and marking. The present invention also discloses a kit which includes the HER-2 gene and / or TOP2A gene detection probe. The best HER-2 gene and TOP2A gene detection probe is screened, so that a signal counting line is accurate and rapid, and results are well reproducible.
Owner:GUANGZHOU LBP MEDICINE SCI & TECH
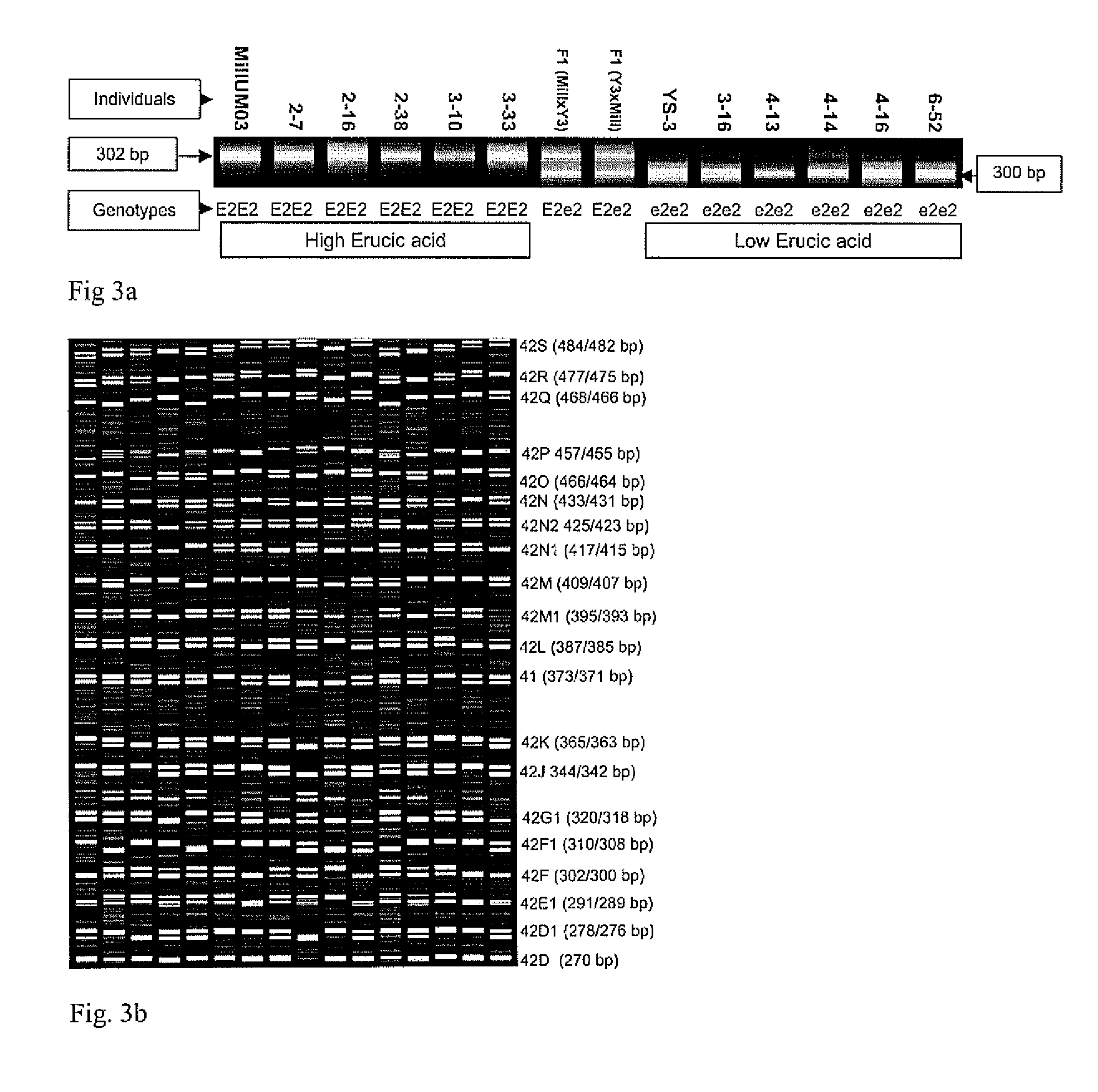
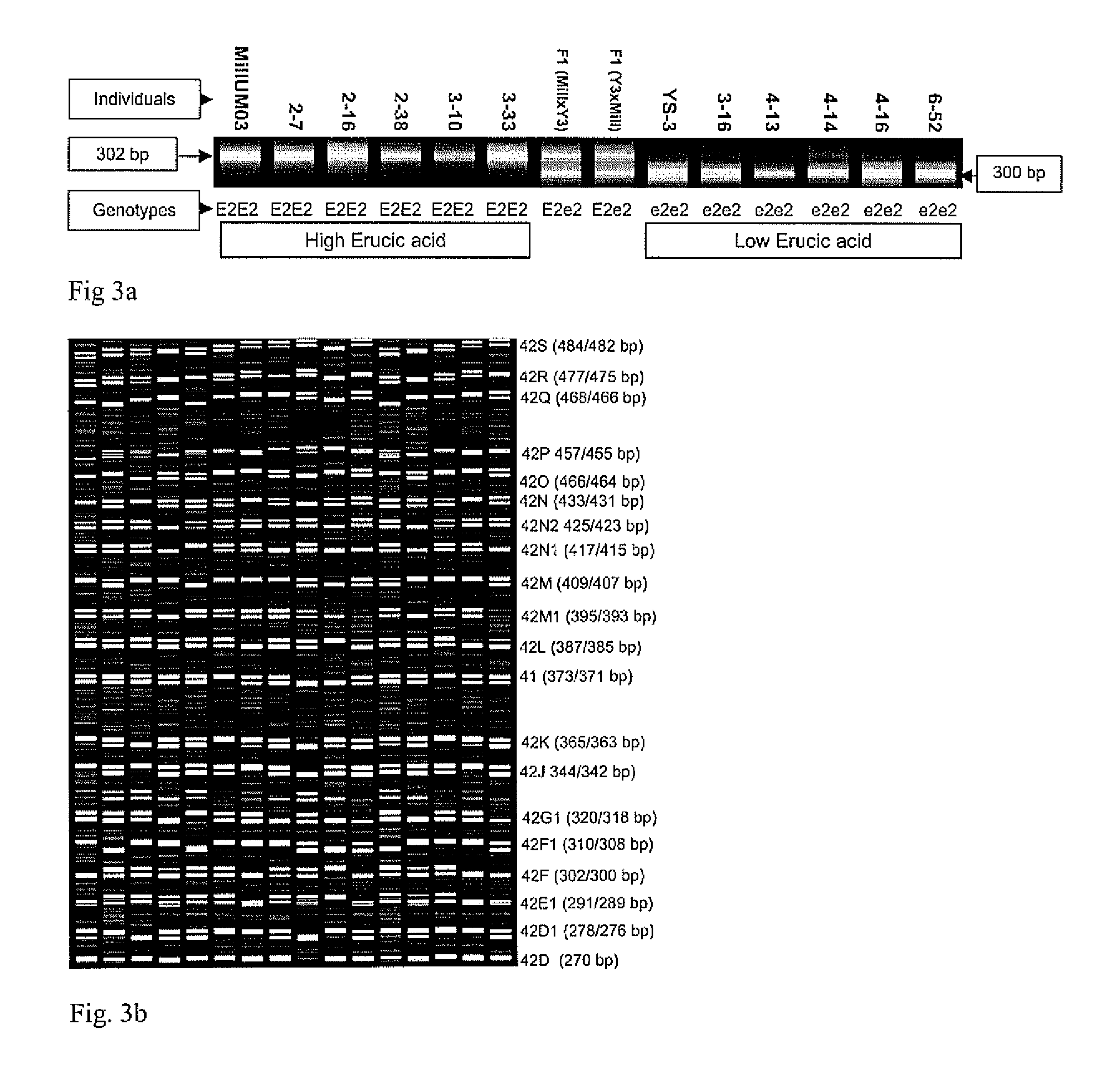
High throughput genome specific molecular markers for erucic acid content genes in brassica napus
InactiveUS20080160530A1Reduce erucic acid contentSugar derivativesMicrobiological testing/measurementBac cloneFluorescence
Owner:LI GENYI
Rapid detection kit and method of NTRK gene fusion
InactiveCN110144401ARapid hybridization testQuick layeringMicrobiological testing/measurementDNA/RNA fragmentationBac clonePolymerase L
The invention relates to a rapid detection method and kit of NTRK gene fusion and belongs to the field of molecular biological detection. According to the rapid detection kit of NTRK gene fusion, a probe is prepared through bought BAC cloning corresponding to the probe by use of phi29DNA polymerase labeling, a buffer solution component is determined by optimization, and the prepared kit can realize rapid hybridization within 2 h and accurate interpretation of NTRK gene rearrangement. The detection kit can better assist existing method for accurate layering, precise diagnosis and treatment of tumor through component optimization and detection of reagent commercialization and has better application prospects.
Owner:GUANGZHOU LBP MEDICINE SCI & TECH
Swine rag-1 gene and utilization thereof
InactiveUS20050155094A1Suppression of gene functionInhibit expressionFungiBacteriaBiotechnologyNucleotide
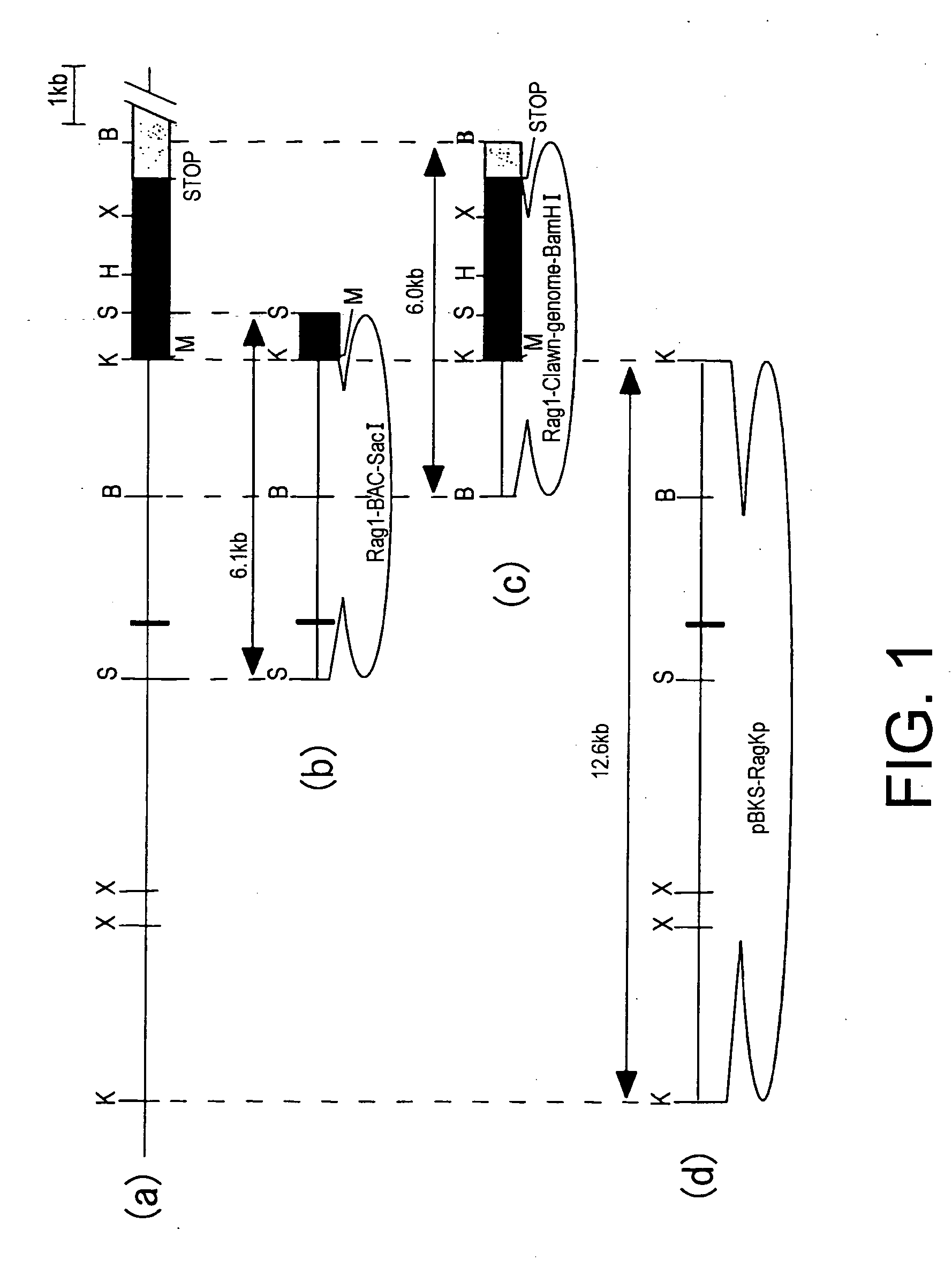
BAC clones comprising the porcine RAG-1 gene were obtained by screening a porcine genomic DNA library. Three clones were obtained by subcloning the region comprising the RAG-1 gene from the BAC clones. Genetic maps were obtained by determining the nucleotide sequence of these clones using sequencing, and by mapping using restriction enzyme digests. Based on this information, targeting vectors that can knockout the RAG-1 gene were constructed. Using these vectors it is possible to construct swine that do not possess acquired immunity by suppressing endogenous porcine RAG-1 gene function.
Owner:NAT INST OF AGROBIOLOGICAL SCI
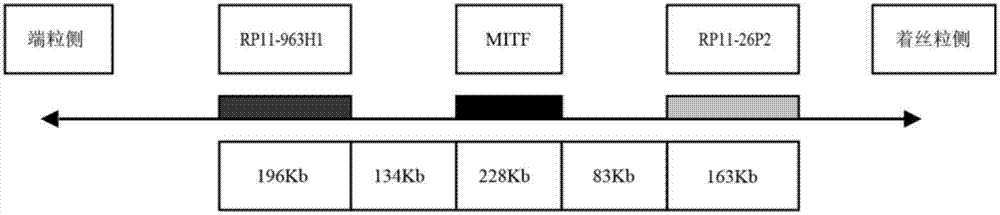
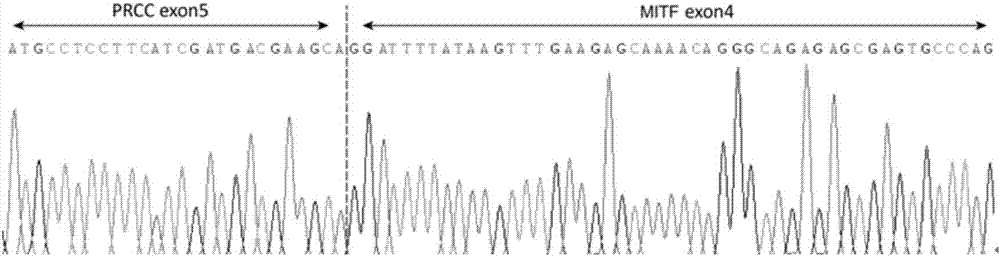
Separating probe combination for diagnosing MITF (Microphthalmia-associated Transcription Factor) translocation kidney cancer and application of separating probe combination
ActiveCN107299152AEasy to detectEasy to separateMicrobiological testing/measurementDNA/RNA fragmentationBac cloneKidney cancer
The invention discloses a separating probe combination for diagnosing MITF (Microphthalmia-associated Transcription Factor) translocation kidney cancer and application of the separating probe combination. The separating probe combination for diagnosing the MITF translocation kidney cancer consists of a BAC (Bacterial Antigen Complex) cloning probe RP11-26P2 and a BAC cloning probe RP11-963H1. According to the separating probe combination disclosed by the invention, a fluorescence labeled DNA (Deoxyribonucleic Acid) combination combined at two ends of an MITF gene is designed according to the characteristics of the MITF translocation kidney cancer; in-situ hybridization, detection fusion and signal separation are carried out on the basis of paraffin embedded tissue sections, so that the accuracy rate of diagnosing tumors can be greatly increased. The probe combination for detecting the MITF translocation kidney cancer, provided by the invention, has the advantages of convenience,` quickness, reliability and high success rate, can be used for preparing a diagnosis kit for the MITF translocation kidney cancer, and provides a novel tool for quickly and accurately diagnosing the MITF translocation kidney cancer.
Owner:NANJING GENERAL HOSPITAL NANJING MILLITARY COMMAND P L A
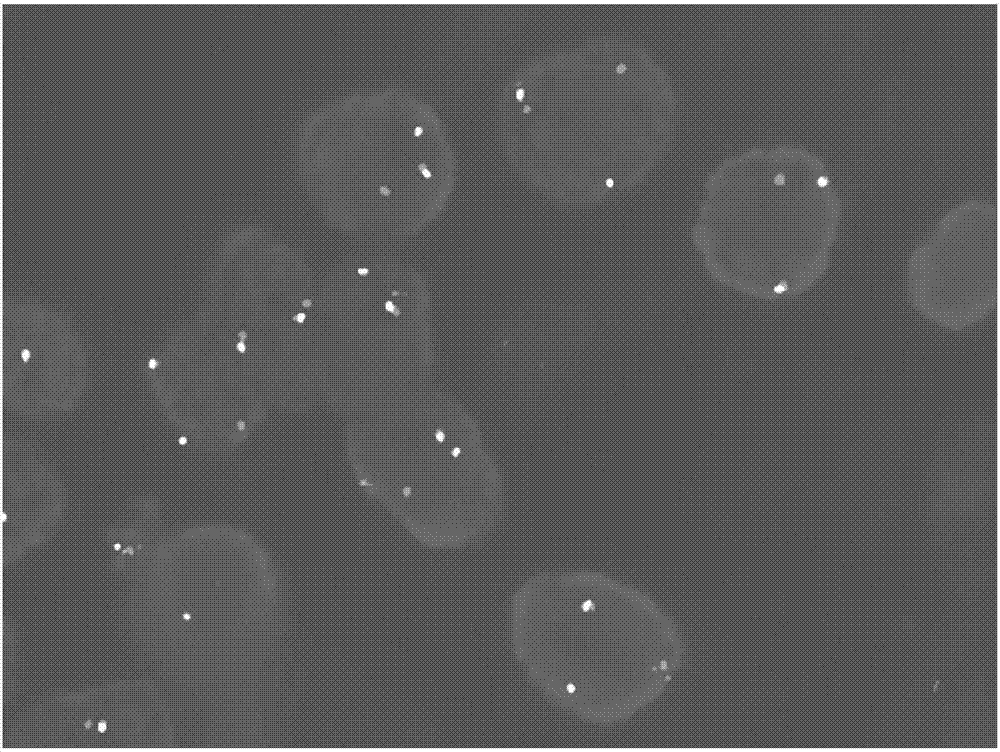
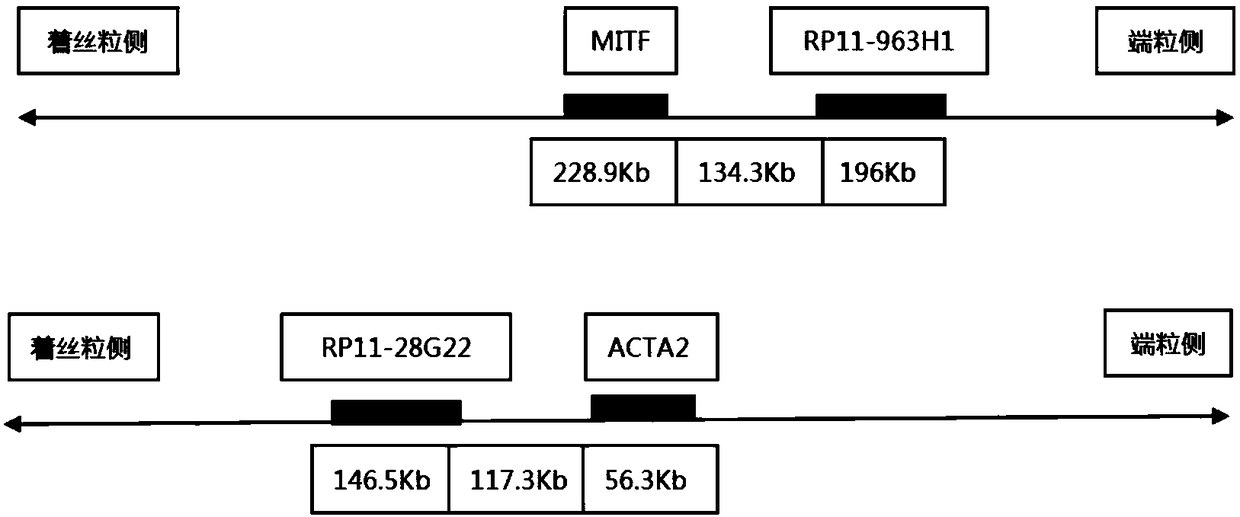
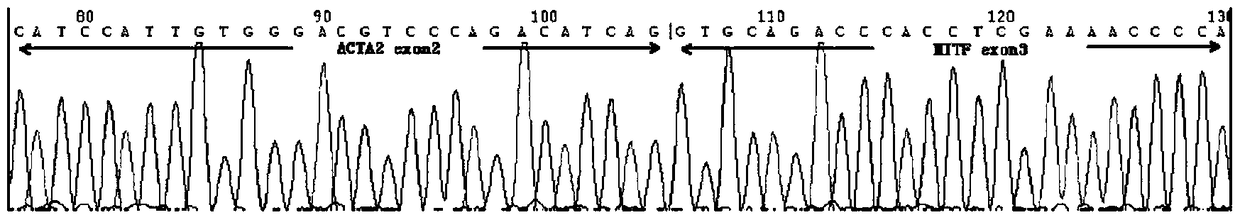
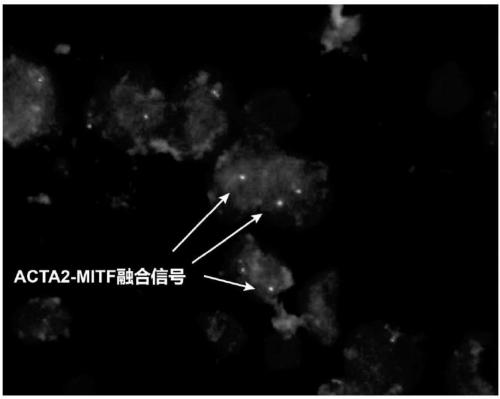
Probe assembly for diagnosing ACTA2-MITF translocation perivascular epithelioid cell tumor and application thereof
ActiveCN109182519AImprove accuracyIncrease success rateMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceWilms' tumor
The invention discloses a probe assembly for diagnosing ACTA2-MITF translocation perivascular epithelioid cell tumor and an application thereof. The probe assembly is formed by a BAC clone probe RP11-28G22 and a BAC clone probe RP11-963H1, wherein the BAC clone probe RP11-28G22 is located on one side of the centromere of ACTA2 and labeled with fluorescence of any color; the BAC clone probe RP11-963H1 is located on one side of the MITF telomere and labeled with fluorescence different from that labeled on the other side of the ACTA2 centromere. As the probe combination carry out in-situ hybridization on the basis of the paraffin-embedded tissue slice to detect the fusion and separation signals, the accuracy rate of diagnosis of such tumors can be greatly improved.
Owner:NANJING GENERAL HOSPITAL NANJING MILLITARY COMMAND P L A
AML1 gene and ETO gene detection probe, preparation method thereof and reagent kit
PendingCN105483253AGood repeatabilityEasy to filterMicrobiological testing/measurementDNA preparationBac clonePlasmid
The invention relates to an AML1 gene and ETO gene detection probe and a preparation method thereof. The method includes the following steps that a selected BAC clone for an AML1 gene is at least one of CTD-3245J1, RP11-77G18, CTD-2349F18, CTD-3171K21 and RP11-384N13, a selected BAC clone for an ETO gene is at least one of RP11-1107H4, RP11-302P1, CTD-2547C5, RP11-55J5 and CTD-2079N17, the DNA of a plasmid is obtained, and marking is conducted. The invention further discloses a reagent kit provided with the AML1 gene and ETO gene detection probe. By obtaining the optimal AML1 gene and ETO detection probe through screening, signal line counting is accurate and quick, and result repeatability is good.
Owner:GUANGZHOU LBP MEDICINE SCI & TECH
Method for animal ubiquitous expression of recombinant human lysozyme
PendingCN106893742ANucleic acid vectorVector-based foreign material introductionBifidobacteriumBac clone
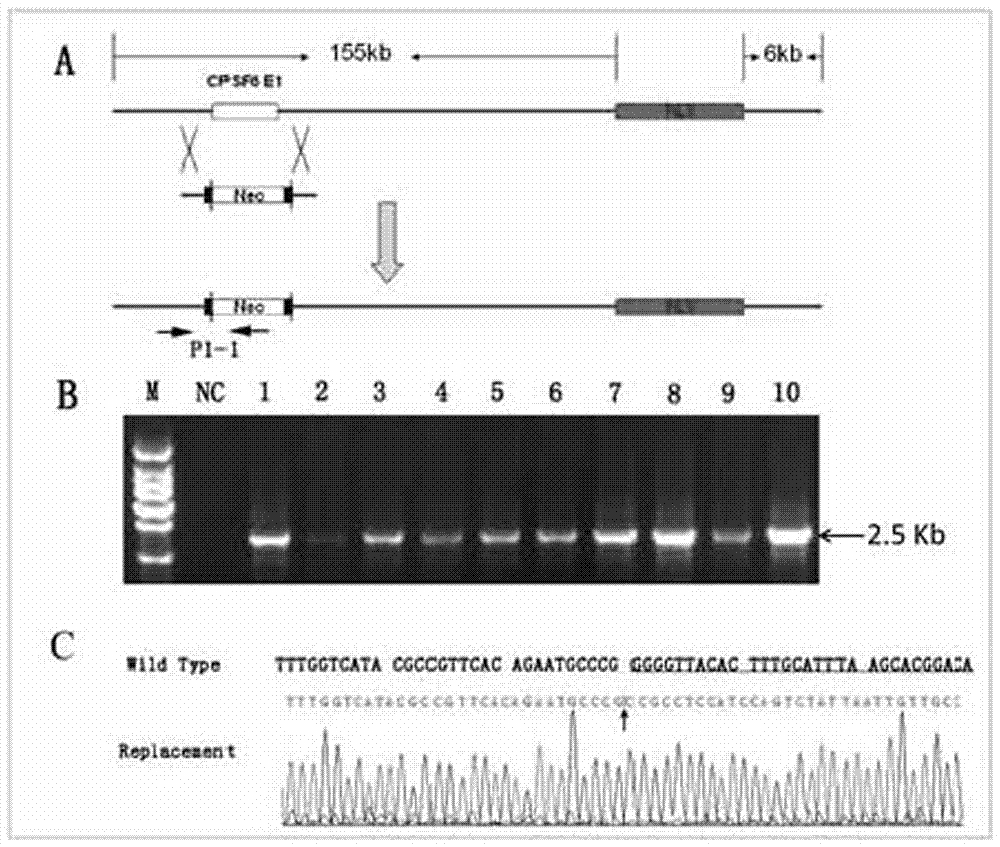
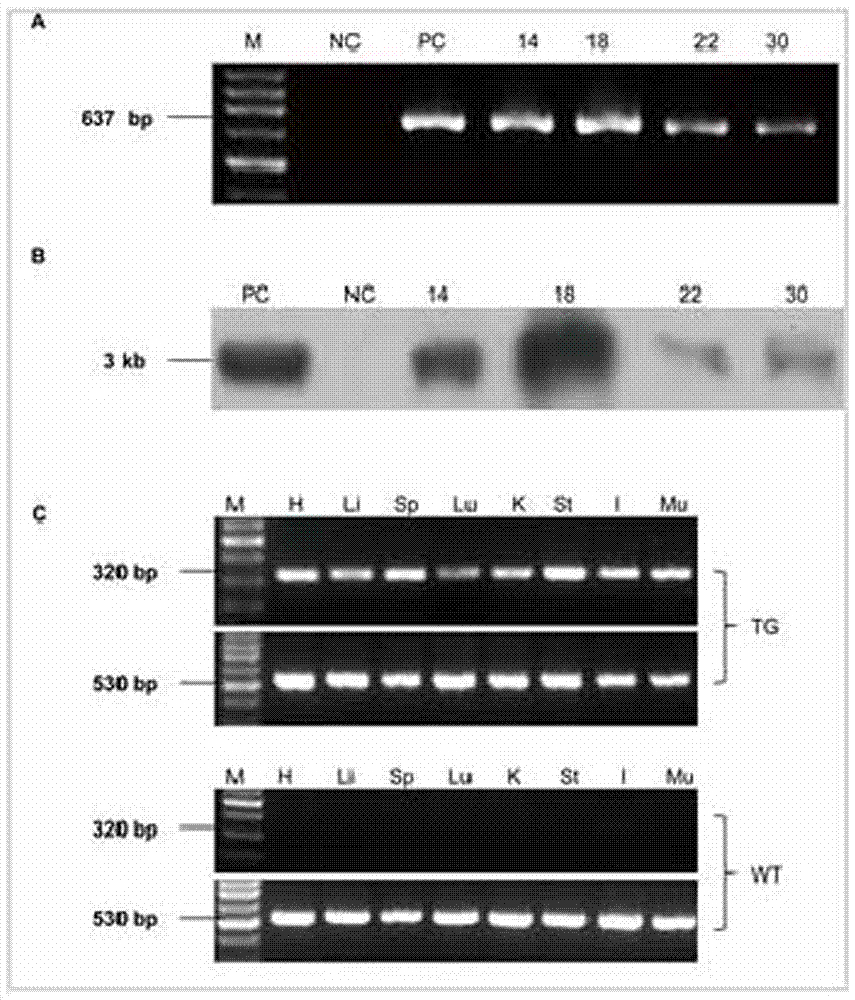
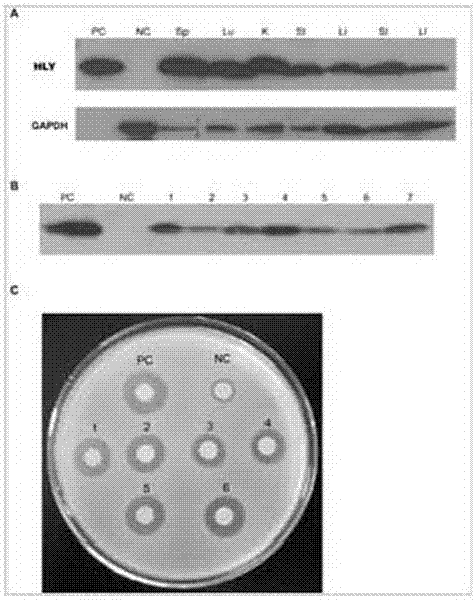
The invention discloses a method for animal ubiquitous expression of recombinant human lysozyme. The method is characterized by comprising the following steps: I, performing human lysozyme BAC cloning, namely, selecting human lysozyme of the Genome Systems company as target hLZ BAC cloning, wherein the GenBank number of the human lysozyme is RP11-72P21; II, removing redundant gene CPSF6, namely, to avoid interference of the redundant gene CPSF6 to the hLZ gene, removing the redundant gene CPSF6 by using a recombinant method, thereby obtaining the human lysozyme pBAC-hLZ-Neo of which the redundant gene is removed. Compared with the prior art, the method has the advantages that the human lysozyme is expressed at multiple parts of bodies. The recombinant human lysozyme expressed in different tissue organs all have bioactivity, the number of bifidobacteria can be remarkably increased (P is less than 0.001) due to the recombinant human lysozyme expressed in intestinal tracts, and meanwhile the number of salmonella can be remarkably reduced (P is less than 0.001).
Owner:FOSHAN UNIVERSITY
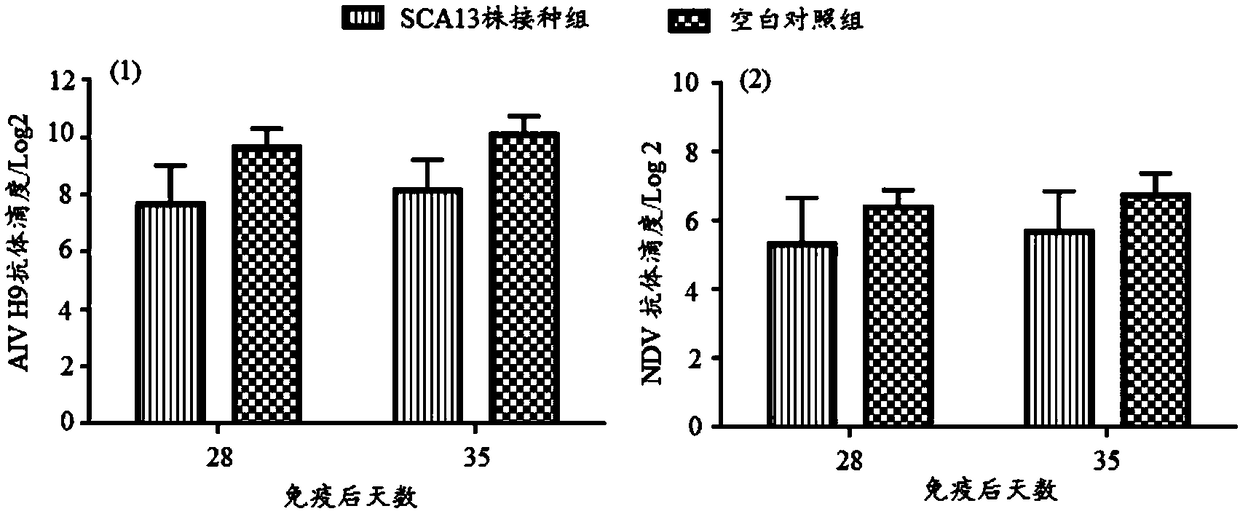
Recombinant Marek's disease virus strain SCA13 strain and application thereof
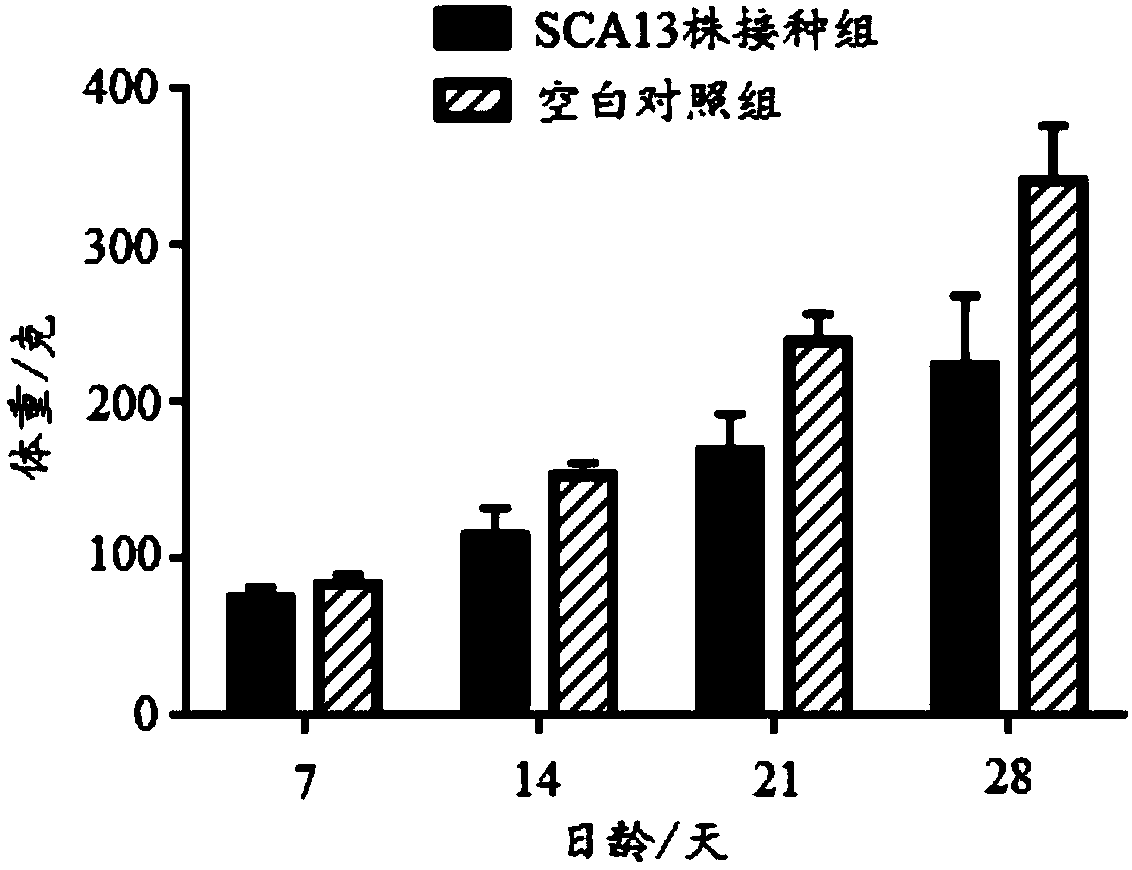
ActiveCN108342367ANon-tumorigenicGood immune protectionViral antigen ingredientsMicrobiological testing/measurementBac cloneHorizontal transmission
The invention discloses a recombinant Marek's disease virus strain SCA13 strain. The biological preservation number of the strain is CGMCC NO.15285, and an SCA13 strain genome is integrated with ALV LTR and can exist stably. On the basis of BAC clone, and by means of the phenomenon that a Red E / T recombinant technology lacks meq and vTR genes, a deletion strain of the meq and vTR genes of the MDVSCA13 strain is built, the biological preservation number of the strain is CGMCC NO.15286, a double gene deletion strain built on the basis of the SCA13 strain completely loses lethality and tumorigenicity to chickens, the immunosuppressive property is lost, the strain has better biosecurity and better horizontal transmission capacity, the problem that in the MD vaccine immunity process, due to immunity leakage of the chickens, diseases occur can be solved, and the chickens are subjected to better immunoprotection in clinic.
Owner:SHANDONG AGRICULTURAL UNIVERSITY
BCR gene and ABL gene detection probe, preparation method thereof and reagent kit
InactiveCN105483256AAchieve identificationFast signal countMicrobiological testing/measurementDNA/RNA fragmentationBCR complexBac clone
The invention relates to a BCR gene and ABL gene detection probe and a preparation method thereof. The method includes the following steps that a selected BAC clone for a BCR gene is at least one of RP11-1026A5, RP11-165G5 and CTD-2079I4, a selected BAC clone is at least one of CTD-2302P22, CTD-2037J11 and CTD-2509L4, a selected BAC clone for an ABL is at least one of CTD-2037L19, RP11-21G10 and CTD-2526G20, the DNA of a plasmid is obtained, and marking is conducted. The invention further discloses a reagent kit provided with the BCR gene and ABL gene detection probe. By obtaining the optimal BCR gene and ABL detection probe through screening, signal line counting is accurate and quick, and result repeatability is good.
Owner:GUANGZHOU LBP MEDICINE SCI & TECH
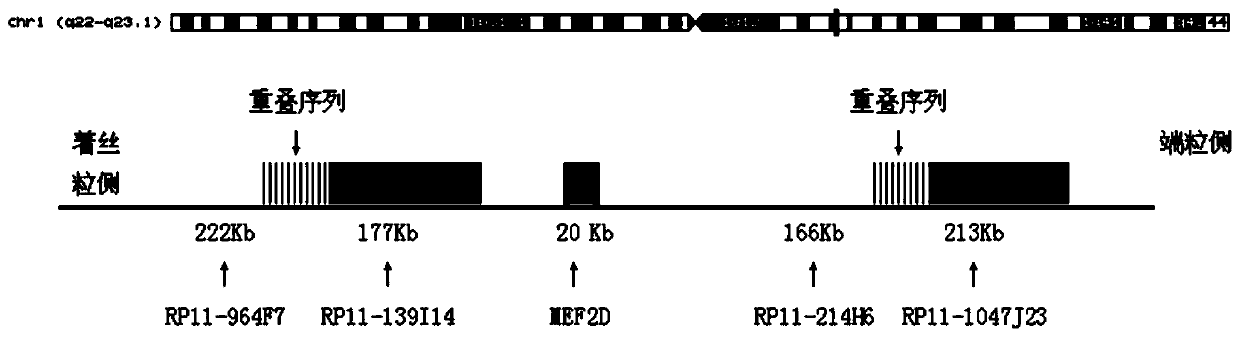
Leukemia MEF2D gene disruption probe detection kit
ActiveCN110093421AGood treatment effectImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationBac cloneFluorescence
The invention discloses a leukemia MEF2D gene disruption probe detection kit. The present invention provides a fluorescent in-situ hybridization polyclonal separating probe for detecting chromosomal MEF2D gene disruption, which is composed of by two BAC clone fragments (RP11-964F7 and RP11-139I14) located on the centromere side of the chromosomal MEF2D gene and two BAC clone fragments (RP11-214H6and RP11-1047J23) located on the telomere side of the chromosomal MEF2D gene. The kit utilizes a FISH technology to detect leukemia associated with MEF2D gene disruption, and performs individualized treatment for patients. The probe of the invention can comprehensively detect all translocations involving the MEF2D gene, finds new translocations, has high application accuracy, high specificity, high success rate, strong fluorescence signal and simple operation, and can assist in the optimization of the treatment and prognosis of leukemia associated with the MEF2D gene disruption.
Owner:BEIJING CHILDRENS HOSPITAL AFFILIATED TO CAPITAL MEDICAL UNIV
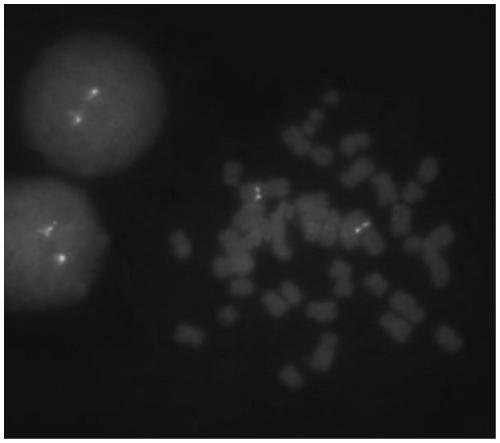
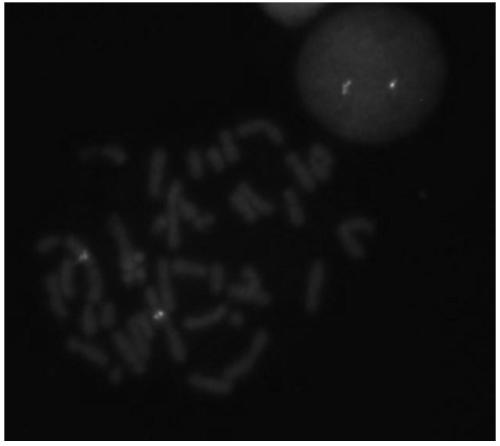
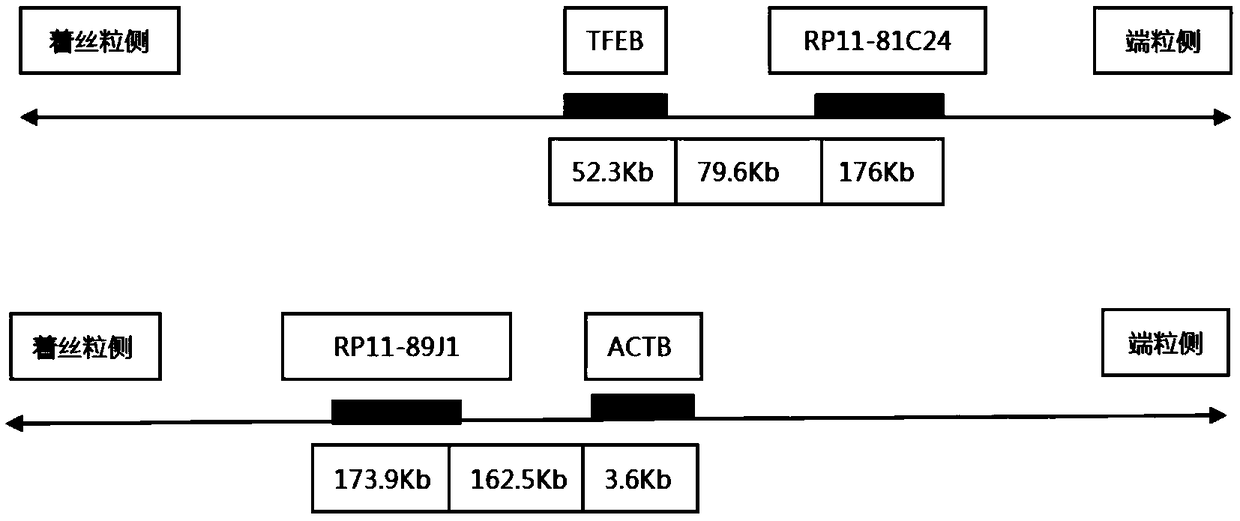
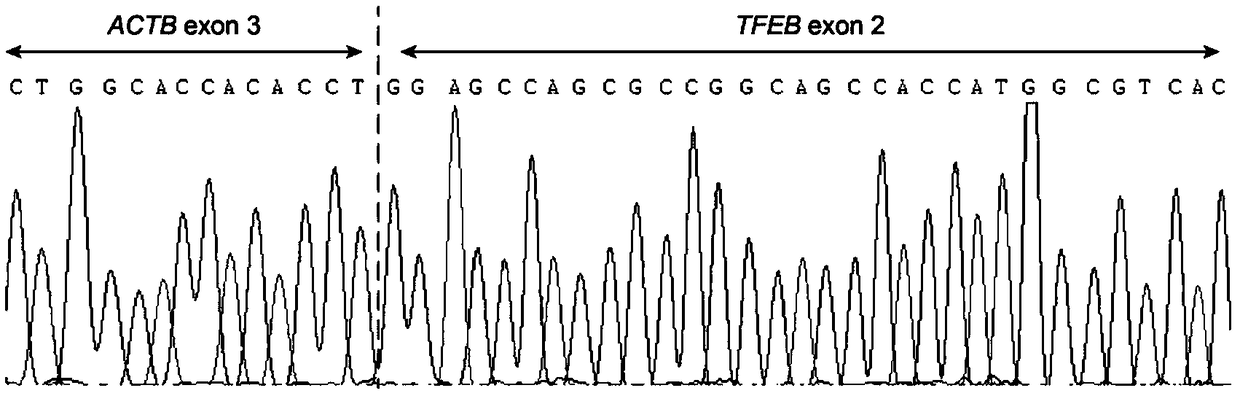
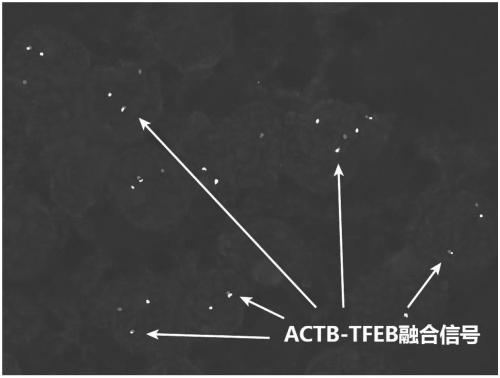
Probe assembly for diagnosing ACTB-TFEB translocation renal carcinoma and application thereof
ActiveCN109022434AImprove accuracyIncrease success rateMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceWilms' tumor
The invention discloses a probe assembly for diagnosing ACTB-TFEB translocation renal carcinoma and application thereof. The probe assembly is formed by a BAC clone probe RP11-89J1 and a BAC clone probe RP11-81C24. The invention discloses application of the probe in preparing an ACTB-TFEB translocation renal carcinoma diagnostic reagent. The invention designs the fluorescence labeling DNA probe assembly combined at the TFEB gene end telomere side and the ACTB gene centromere side according to the characteristic of the TFEB translocation renal carcinoma, in-situ hybridization is carried out onthe basis of a paraffin-embedded tissue slice, and fusion and separation signals are detected, so that the accuracy on diagnosing the tumors can be greatly improved.
Owner:NANJING GENERAL HOSPITAL NANJING MILLITARY COMMAND P L A
Methods for preparing gene chips and use thereof
InactiveUS20070166714A1Easy to copyBioreactor/fermenter combinationsBiological substance pretreatmentsPolynucleotideBiology
The invention relates to methods and compositions for producing biochips as well as to the use of these biochips in diverse fields, from functional genomics to diagnosis, for example, particularly in research or in the medical field. The invention also relates to tools and methods for selecting polynucleotides that permit the production of improved biochips. The inventive biochips contain, in particular, BAC clones.
Owner:INTEGRAGEN
Breeding method for transgenic mouse with conditional knockout of lncRNA DLX6-os1
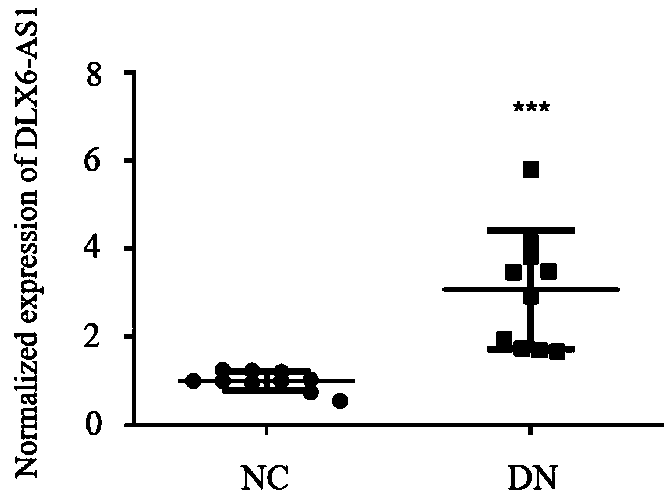
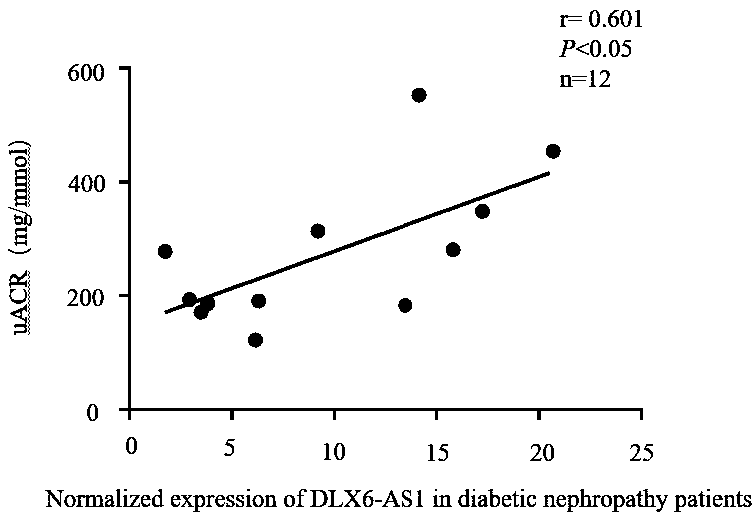
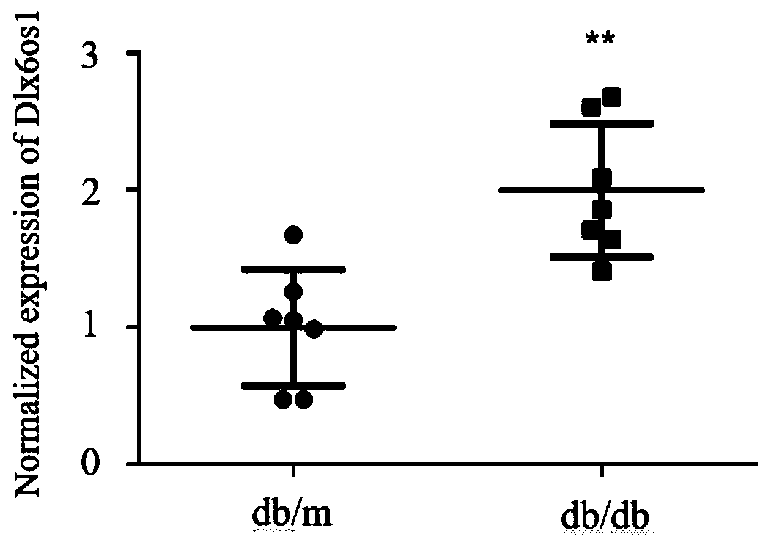
PendingCN110117616AOpen up new avenuesStrong use valueStable introduction of DNANucleic acid vectorExonConditional gene knockout
The invention relates to a breeding method for a transgenic mouse with conditional knockout of lncRNA DLX6-os1. The purpose is effectively achieved that the transgenic mouse as an animal model adaptsto different tissue organs for studying the effects of the lncRNA. A Dlx6-os1 conditional knockout carrier, the Dlx6-os1 gene, a transcript Dlx6-os1-201 ENSMUST00000159568.5 and three exons located ata chromosome 6 of the mouse are constructed, and the three exons are used as a conditional knockout area; a BAC clone of RP24-276P7 and RP24-260F14 in a C57BL / 6 mouse gene library is used as a template for PCR operation; in the purpose carrier, a self-deleting anchoring locus is inserted beside an Neo locus, an loxP locus is inserted beside the gene knockout area, and DTA negative selection is conducted; a gene mediated with a Cre enzyme is recombined, and a gene knockout plasmid carrier is obtained; the gene knockout plasmid carrier is transferred into an ES cell and hybridized with multipletransgenic mice with the Cre enzyme in different organs, and positive mice with the reduced expression level of the DLX6-os1 in different organs are obtained. The method is suitable for studying theeffects of the lnc RNA in different tissue organs, and a new way for diabetes diagnosis and treatment is developed.
Owner:THE FIRST AFFILIATED HOSPITAL OF ZHENGZHOU UNIV
TOP2A gene detection probe, preparation method thereof and reagent kit
InactiveCN105483255AStrong specificityHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationTOP2A GenePlasmid dna
The invention relates to a TOP2A gene detection probe and a preparation method thereof. The method includes the following steps that a selected BAC clone is at least one of RP11-1152A10 and CTD-3217C5, or a selected BAC clone is at least one of RP11-737D6, CTD-3087O22, RP11-48O10 and CTD-2134K5, a plasmid is extracted from the clone to obtain DNA of the plasmid, and quantification is conducted; marking is carried out with fluorescein. The invention further discloses a reagent kit provided with the TOP2A gene detection probe. By obtaining the optimal TOP2A detection probe through screening, signal line counting is accurate and quick, and result repeatability is good. Defects of TOP2A mutation detection in clinic are made up for, more patients benefited from targeted drug can be screened more easily, the survival rate of the patients is increased, and overall life time is prolonged.
Owner:GUANGZHOU LBP MEDICINE SCI & TECH
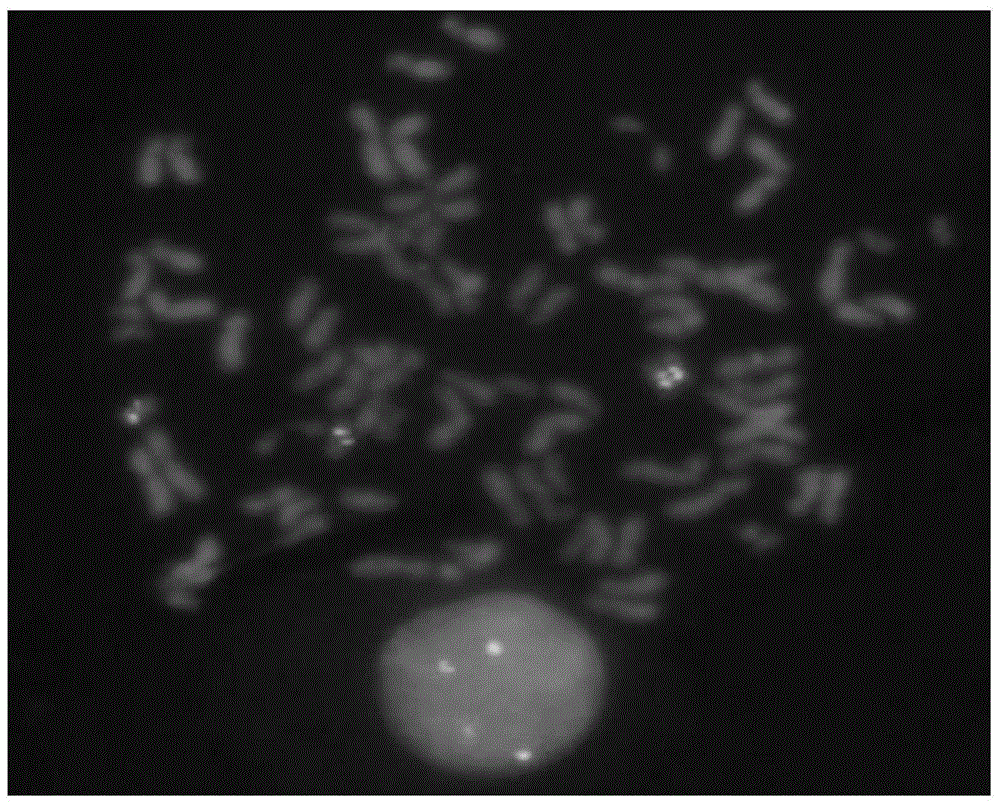
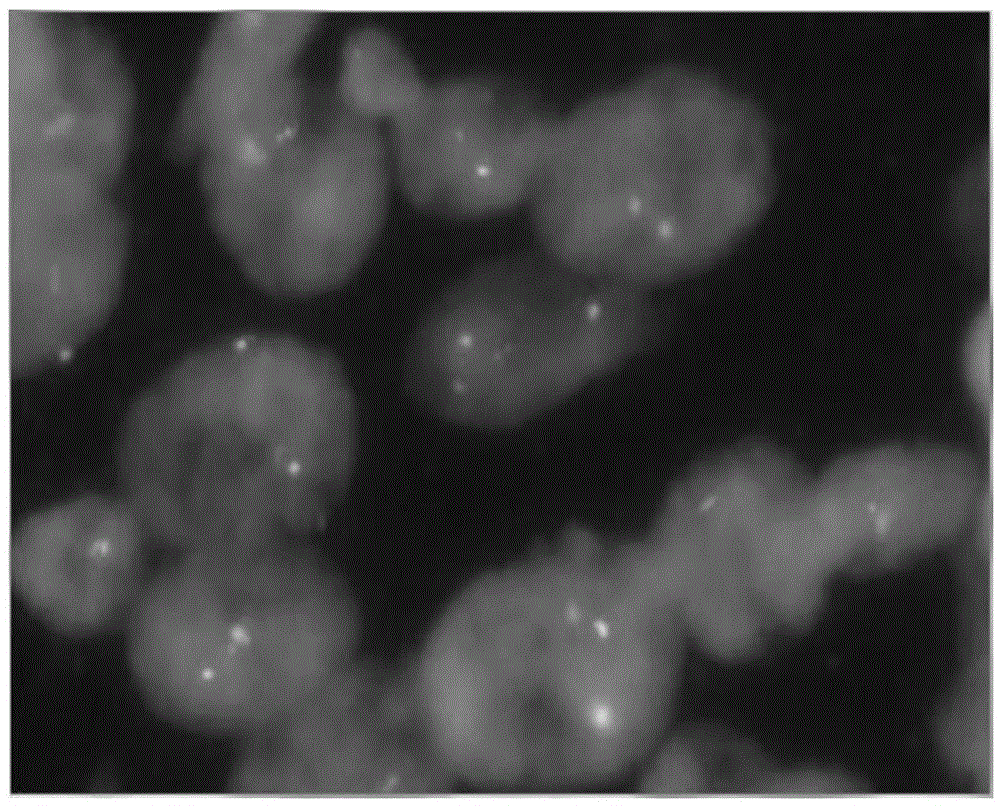
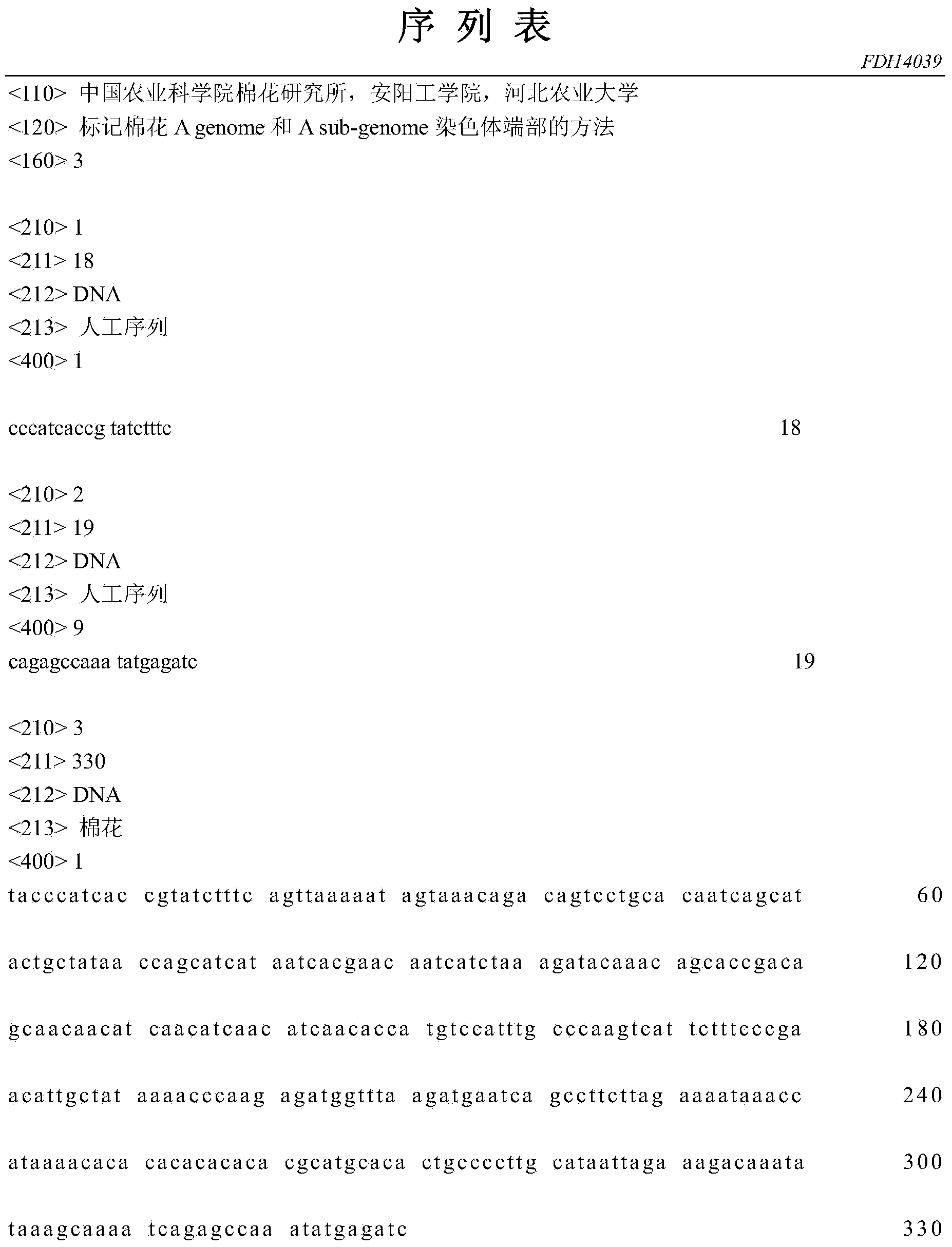
Method for marking chromosome terminals of cotton A genome and A sub-genome
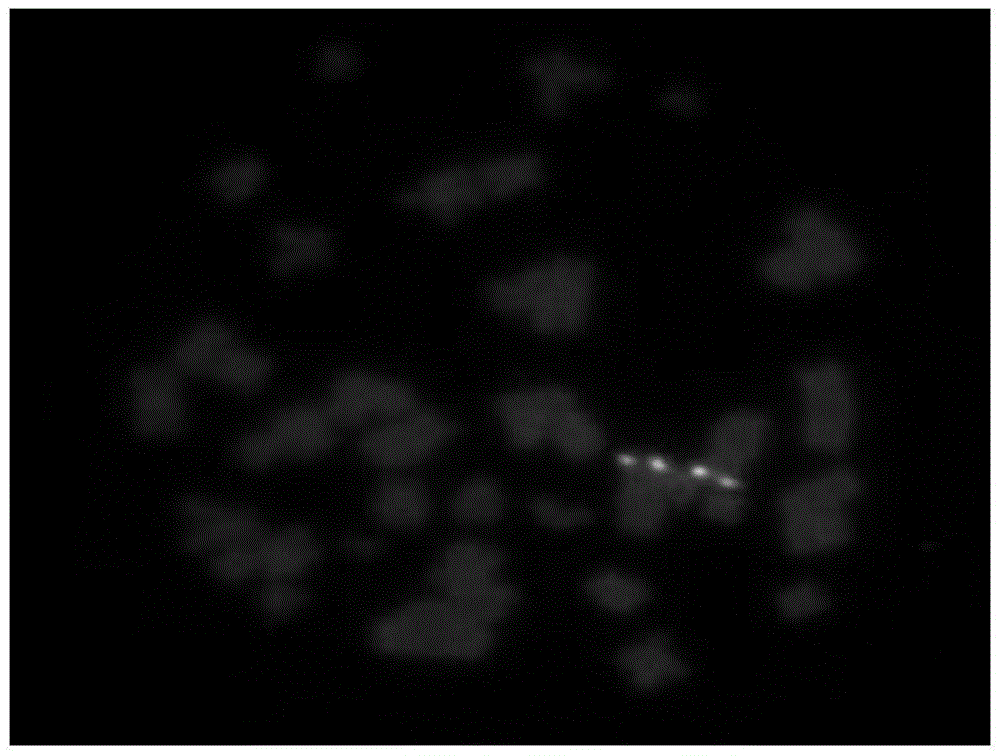
The invention belongs to the field of molecular cytogenetic, and concretely relates to a method for marking chromosome terminals of cotton A genome and A sub-genome. The method comprises: using BAC coming from gossypium barbadense pima-90BAC library to clone 350B21, performing BAC-FISH on mitosis mid-term chromosomes of different cotton species, and discovering the phenomena that the terminals of all chromosomes of A genome and A sub-genome generate strong hybridization signals, while the hybridization signals of all chromosomes of D genome and D sub-genome are not obvious. By utilizing the BAC-FISH method for BAC cloning, the chromosome terminals of cotton A genome and A sub-genome can be rapidly effectively marked.
Owner:INST OF COTTON RES CHINESE ACAD OF AGRI SCI +2
Promoter for heat shock protein gene of litopenaeus vannamei
InactiveCN102757964AEasy accessHigh biosecurityVector-based foreign material introductionDNA/RNA fragmentationFluorescenceTarget gene
The invention relates to the biotechnical field, in particular to a promoter for a heat shock protein gene of litopenaeus vannamei. The promoter for the heat shock protein gene of the litopenaeus vannamei is obtained by bacterial artificial chromosome (BAC) clone screening and sequencing analysis, promoter prediction, DNA amplification and sequencing, vector construction and other technologies, and has the base sequence at the sequence table SEQ ID NO.1; then, the cloned promoter is combined with a plasmid carrying an enhanced green fluorescence protein (EGFP) to construct an EGFP gene expression vector containing a litopenaeus vannamei autochthonous promoter; and the expression vector is transfected to an insect cell sf9 to observe that the green fluorescence protein has a strong expression and can have a higher expression after heat shock. The promoter for the heat shock protein gene of the litopenaeus vanname is obtained by a clone full sequencing method for the BAC containing a target gene and the promoter prediction method, which are more simple and fast than the conventional chromosome walking method.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Rice aroma gene of paddy rice and constructing process and use of expression vector including the rice aroma gene
InactiveCN1810974ABreed fastFermentationVector-based foreign material introductionStart codonBac clone
The present invention relates to rice breeding gene technology in biotechnology field, and is rice aroma gene of paddy rice and the constructing process and use of expression vector including the rice aroma gene. The rice aroma gene of paddy rice has nucleotide of total length of 9066 bases and including 1242 bases in start codon ATG upstream, 5859 bases in the gene region and 1965 bases in the termination codon TAA downstream, coding area comprising 15 exons, and gene sequence as changed betaine aldehyde dehydrogenase coding gene Badh with an 8 base deletion in the 7-th exon. Through cutting the integral rice aroma gene of paddy rice of the positive BAC clone with the limiting cleavage sites HindIII on two sides of the rice aroma gene of paddy rice Badh and cloning to expression vector pCAMBIA1302, the expression vector of rice aroma gene of paddy rice may be obtained and may be used in breeding excellent rice variety.
Owner:YANGZHOU UNIV
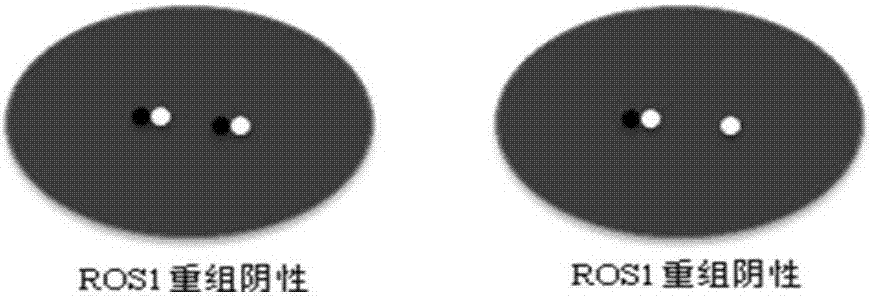
Kit and detection method for abnormity of ROS1 and C-met genes
InactiveCN107418999AStrong specificityReduce background noiseMicrobiological testing/measurementBac cloneConserved sequence
The invention discloses a kit and a detection method for abnormity of ROS1 and C-met genes. The kit includes a first group of probes and a second group of probes, which aim to the ROS1 gene, and / or a third group of probes and a fourth group of probes, which aim to the C-met gene. Each group of probes is labeled with a fluorescent dye, wherein the color of the fluorescent dye on the probes in the same group is same while the colors of the fluorescent dyes on the probes in different groups are different. The four groups of probes respectively are amplification products by amplifying a primer with corresponding BAC cloning as a template. The FISH probes are produced through repeated comparison and selection and then utilization of optimized BAC cloning as the template and through amplification by designing a primer aiming to a non-repeated and highly-conserved sequence, so that the kit has excellent specificity and lower background noise. The kit can achieve the optimal balance between detection specificity and hybridization time length, so that not only are specificity and sensitivity of a result ensured, but also hybridization time is reduced, thereby increasing detection efficiency.
Owner:SUREXAM BIO TECH
Reagent for detecting copy number of EGFR gene and ploidy of chromosome 7
ActiveCN101899504BQuick checkEasy to detectMicrobiological testing/measurementFluorescence/phosphorescenceTyrosine-kinase inhibitorPloidy
Owner:合肥艾迪康医学检验实验室有限公司
White flower beet the 9th chromosome BAC chip and its preparation method
InactiveCN1570144AQuick analysisEfficient analysisMicrobiological testing/measurementGene mappingBac clone
A white bloom beet chromosome 9 BAC chip and its preparing method are provided. BAC clone on the BAC chip is obtained by screening the BAC library using white bloom beet chromosome, and the chip is prepared by combining the BAC clone with the carrier. The preparing method comprises: primary treatment for BAC, BAC treatment before spotting, and preparation of the BAC chip. M14 and M14 chromosome addition line differential expression gene screening, genome differential analysis, gene mapping and number of replications analysis can be done rapidly and efficiently by using the chip prepared by the method.
Owner:HEILONGJIANG UNIV +1
ALK gene and EML4 gene detection probe, preparing method thereof and kit
InactiveCN105543353AGood repeatabilityAccurate signal countMicrobiological testing/measurementDNA preparationBac clonePlasmid dna
The invention relates to an ALK gene detection probe, an ALK gene and EML4 gene detection probe and a preparing method thereof. The preparing method comprises the following steps that BAC aiming at an ALK gene is selected to be cloned into at least one of RP11-62B19, RP11-100C1 and CTD-2245E6, and BAC is selected to be cloned into at least one of CTD-2544I11, RP11-684O3 and RP11-134O13; BAC aiming at an EML4 gene is selected to be cloned into at least one of CTD-2358L8 and CTD-2547J15; plasmid DNA is obtained; marking is carried out. The invention further discloses a kit containing the ALK gene and EML4 gene detection probe. The optimal ALK gene and EML4 gene detection probe is obtained by means of screening, signal counting is accurate and fast, and result repeatability is good.
Owner:GUANGZHOU LBP MEDICINE SCI & TECH
High throughput genome specific molecular markers for erucic acid content genes in Brassica napus
A single base change in the Bn-FAE1.1 gene in the A genome and a two-base deletion in the Bn-FAE1.2 gene in the C genome produce the nearly zero content of erucic acid observed in canola. A BAC clone anchoring Bn-FAE1.1 from a B. rapa BAC library and a BAC clone anchoring Bn-FAE1.2 from a B. oleracea BAC library were used in this research. After sequencing the gene flanking regions, it was found that the dissimilarity of the flanking sequences of these two FAE1 homologs facilitated the design of genome specific primers that could amplify the corresponding genome in allotetraploid B. napus. The two-base deletion in the C genome gene was detected as a sequence characterized sequence region (SCAR) marker. To increase the throughput, one genome specific primer was labeled with four fluorescence dyes and combined with 20 different primers to produce PCR products with different fragment sizes. Eventually, a super pool of 80 samples was detected simultaneously, making it possible to analyze over half a million of samples per day using a medium capacity ABI 3100 Genetic Analyzer. This dramatically reduces the cost of marker detection. The single base change in the Bn-FAE1.1 gene was detected as single nucleotide polymorphic (SNP) marker with an ABI SNaPshot kit. A multiplexing primer set was designed by adding a polyT to the 5′ primer end to increase SNP detection throughput through sample pooling. These multiplexed high throughput molecular markers have been successfully implemented in our canola / rapeseed breeding programs.
Owner:LI GENYI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com