Probe assembly for diagnosing ACTA2-MITF translocation perivascular epithelioid cell tumor and application thereof
An ACTA2-MITF and perivascular technology, which is applied in the field of fluorescence in situ hybridization probes, can solve the problem of no report on the detection of ACTA2-MITF fusion genes, achieve reliable success rate, and improve the effect of accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1 is verified for the case of definite diagnosis:
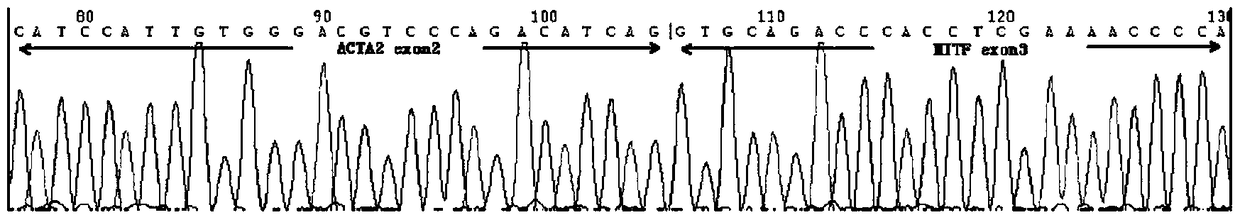
[0038] Through high-throughput sequencing technology, the team of the present invention detected the ACTA2-MITF fusion gene in a case whose morphology conformed to the characteristics of perivascular epithelioid cell tumors, and found that the gene was formed by the fusion of ACTA2 exon 2 and MITF exon 3 . For this case in which high-throughput sequencing RNA-seq detected a new fusion gene of ACTA2 exon2-MITF exon3, PCR RT-PCR was performed on this case using the primers we designed.
[0039] 1. Extraction of RNA:
[0040] Strictly follow the RNeasy FFPE Kit operating instructions for extraction. ① Dewaxing: dewax the collected slides with xylene, rinse with absolute ethanol, air-dry and scrape off with a scalpel blade and put them into a 1.5ml EP tube; Proteinase K, mix well, enzymatically digest at 56°C for 15min, then at 80°C for 15min, cool on ice; ③add 16μl DNase buffer, then add 10μl DNase I, mix we...
Embodiment 2
[0044] Embodiment 2: the preparation of DNA probe combination:
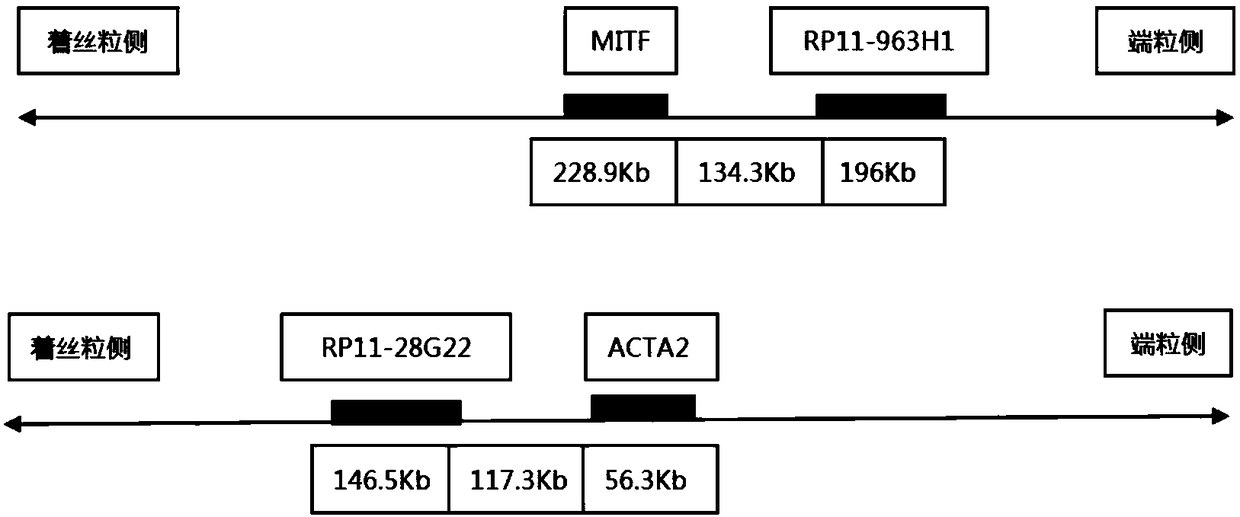
[0045] Select two BAC clone fragments that can be connected at the telomeric side of the MITF gene on chromosome 3 and the centromere side of the ACTA2 gene on chromosome 10, and control the farthest distance between the probes at both ends within 1500kb, and keep the distance between the BAC clone fragments A certain distance does not overlap, and the fragments are similar in size. The cloned fragments were obtained from the Human BAC Cloning Center of EmpireGenomics (http: / / www.empiregenomics.com / helixhq / clonecentral / search / human). The BAC clone probe on the centromere side of ACTA2 is RP11-28G22 (fragment length 146.5kb), and the BAC clone probe on the telomeric side of MITF is RP11-963H1 (fragment length 196kb). These fragments are Bacterial artificial chromosomes, BAC) clone, its location on human chromosomes has been published, respectively, RP11-28G22 is located on chromosome 10 90430957-90577498, and RP1...
Embodiment 3
[0047] Embodiment 3: fluorescence in situ hybridization process:
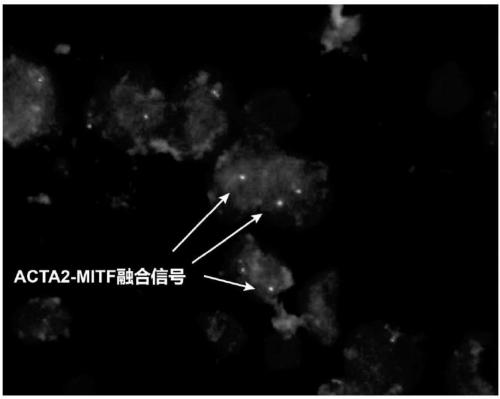
[0048] One case of ACTA2-MITF translocation PEComa was confirmed by high-throughput sequencing and RT-PCR detection of fusion genes ( figure 2 , corresponding to the results of this experiment can better reflect the reliability of this experiment), and two experienced pathologists refer to the WHO 2016 urinary and male reproductive system classification standards to collect renal cell carcinoma and PEComa in Nanjing General Hospital of Nanjing Military Region. A total of 30 cases served as the control group.
[0049] Wax blocks were sliced at a thickness of 3 μm. After dewaxing, they were placed in 100%, 85%, and 70% ethanol for 2 minutes each, and then immersed in deionized water for 15 minutes in a water bath at 100°C. Put the tissue slices into pepsin K solution (0.1g pepsin, 40ml 0.01M HCL) at 37°C for 15min; rinse twice with 2×SSC (sodium chloride, sodium citrate) for 5min each, and place the slices in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com