Chlorinated aliphatic hydrocarbon degradative plasmid pRC11, engineered bacteria and application thereof
A technology of chlorinated aliphatic hydrocarbons and PRC11, which is applied to the separation of bacteria, dispersed particles, and air quality improvement. It can solve the problems of no reports of chlorinated aliphatic hydrocarbons degrading plasmids and late organic pollutants degrading plasmids, so as to improve biological Degradation effect, improvement of processing capacity, effect of clear genetic background
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
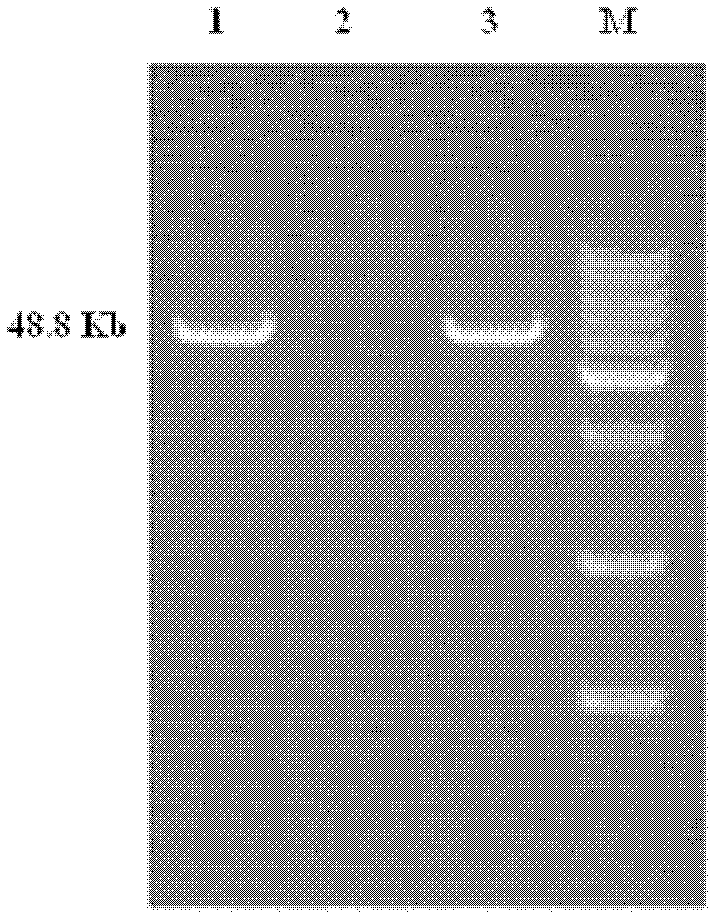
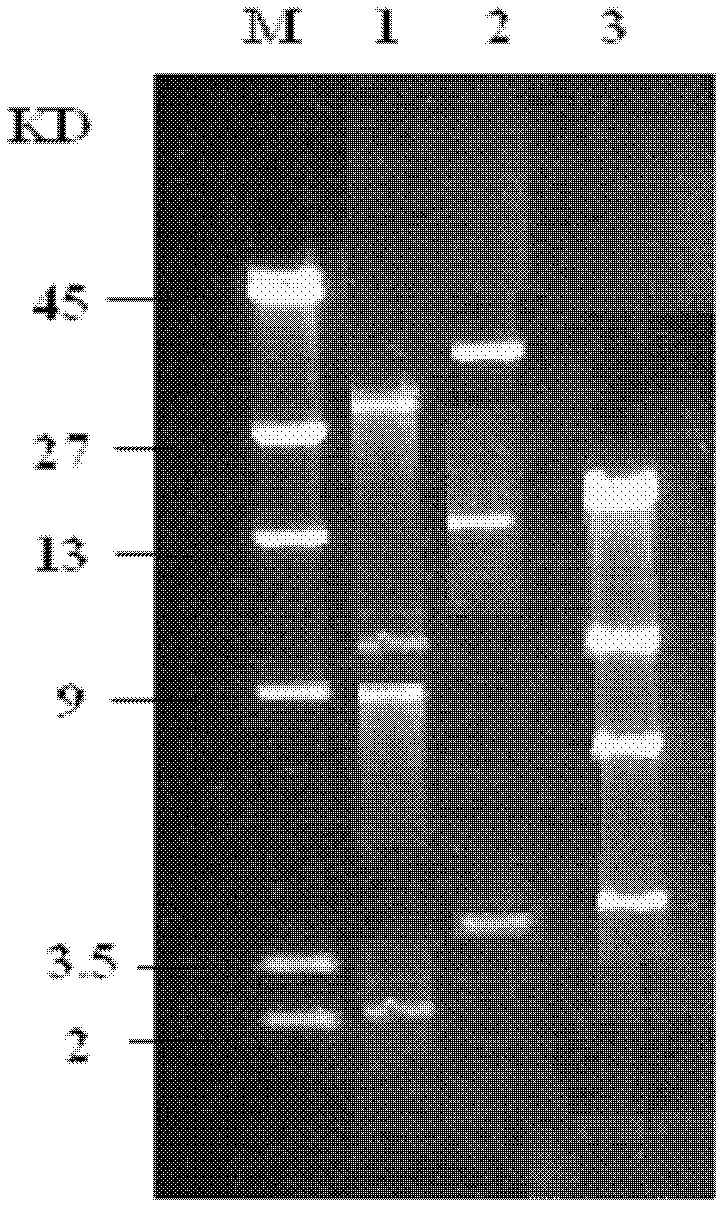
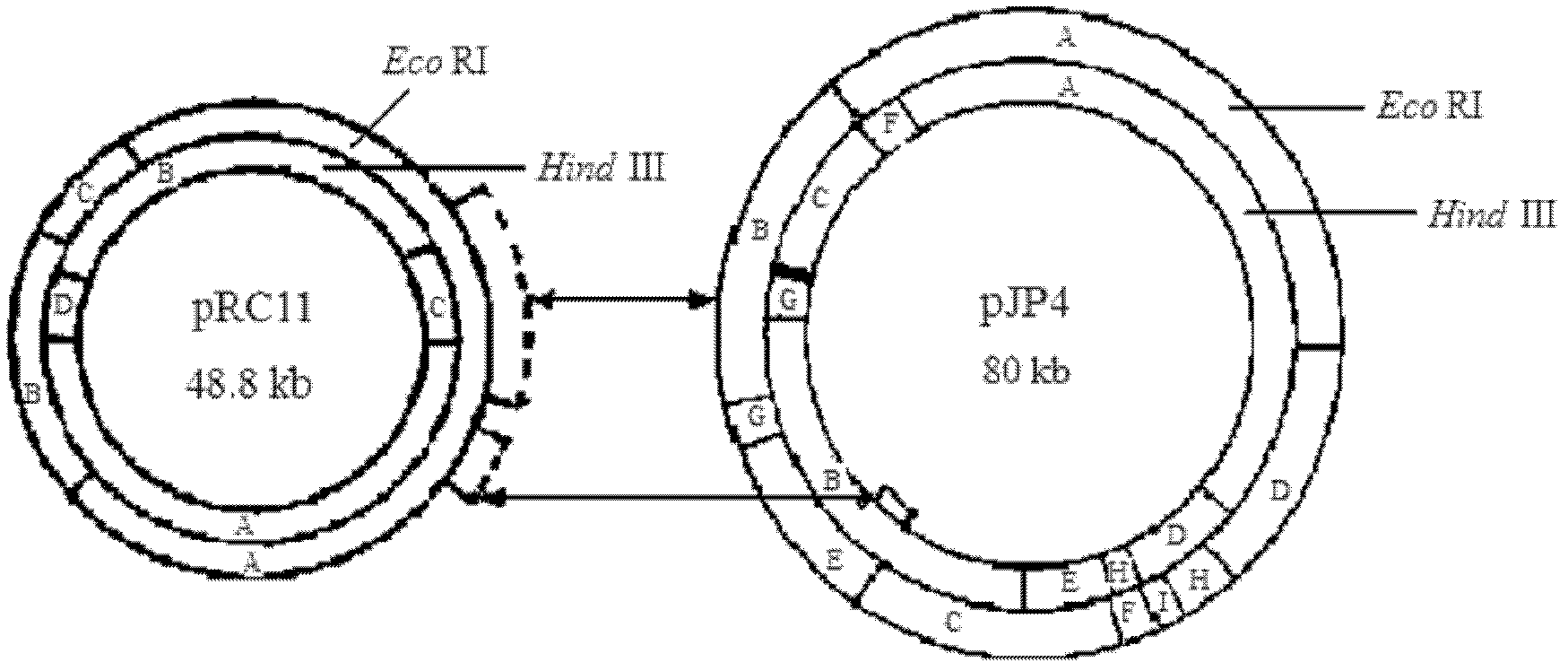
[0019] Example 1: Acquisition and Identification of Plasmid pRC11
[0020] 1) Plasmid detection:
[0021] (1) Cultivate 3 mL of LB bacterial solution overnight, centrifuge at 7000 rpm at 4°C for 10 minutes;
[0022] (2) Resuspend in 1 mLLTE buffer (40mM Tris-acetic acid, 2mM EDTA, adjust the pH value to 7.9 with glacial acetic acid);
[0023] (3) Add 2mL of lysate (3% SDS, 50mM Tris, add 1.6mL of 2N NaOH to adjust the pH to 12.6) for lysis, and shake gently to mix:
[0024] (4) Heating in a water bath at 50-65°C for 20 minutes, then adding 2 times the volume of chloroform;
[0025] (5) Gently oscillate to emulsify, and centrifuge to break the emulsion (6000 rpm, 15 minutes, 40°C);
[0026] (6) The supernatant was directly analyzed by electrophoresis.
[0027] Plasmid bands were analyzed by agarose gel electrophoresis under the following conditions:
[0028] Agarose concentration 0.7%, electrophoresis voltage 80V, electrophoresis buffer TBE, marker is λDNA / HindIII, adopt g...
Embodiment 2
[0055] Example 2: Transformation of plasmid E.coli DH5
[0056] (1) Transformation and amplification of plasmids:
[0057] Preparation of Competent Cells: Use an inoculation loop to pick out the DH5α strains preserved in glycerol and streak them on LB agar plates. Take a single colony on the agar plate, inoculate it into 5mL LB medium, and cultivate it with shaking at 37°C until the OD 600 = 0.3-0.4, cool the culture tube to 0-4°C. Take several sterilized 1.5mL centrifuge tubes and add 200μL of bacterial solution to each. Then add filter-sterilized 4mol / L CaCl 2 5 μL, capped, and flicked the bottom of the tube to mix. Competent bacteria were obtained.
[0058] Plasmid transformation: Add 5 μL of plasmid solution (DNA≤10ng) to the competent cells, flick the bottom of the centrifuge tube to mix the plasmid and bacteria evenly. Stand in an ice bath at 0-4°C for 30 minutes. In a 42°C water bath, place quietly for 90-120s. Ice bath 1 ~ 2min. Incubate with shaking at 37°C f...
Embodiment 3
[0061] Embodiment 3: Plasmid pRC11 positive transformant E.coli DH5 (dcm + ) in the application of biological filtration treatment of dichloromethane
[0062] (1) Bacterial strain culture: adopt carbon-free Chapei solid medium for cultivation, first coat the strains on the surface of the solid medium, then add a large piece of sterile cotton on the lid of the dish, drop an appropriate amount of dichloromethane in the cotton, place Cultivate at 30°C, and add dichloromethane regularly. Add sterile physiological saline to the cotton on the control plate, and after cultivating for 2 to 3 days, the single colony obtained is transferred to the slant of the large test tube, and cotton dripped with dichloromethane is placed on the opposite side of the slant to obtain the slant bacteria. kind.
[0063] (2) Bacterial strain expansion culture: the slant bacterial strain is cultivated in the seed culture medium to obtain the bacterial suspension, 50ml bacterial suspension (cell concentr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com