Patents
Literature
247 results about "Epigenetic Profile" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
The analysis of all of the epigenetic DNA modifications in the genome a biological sample.
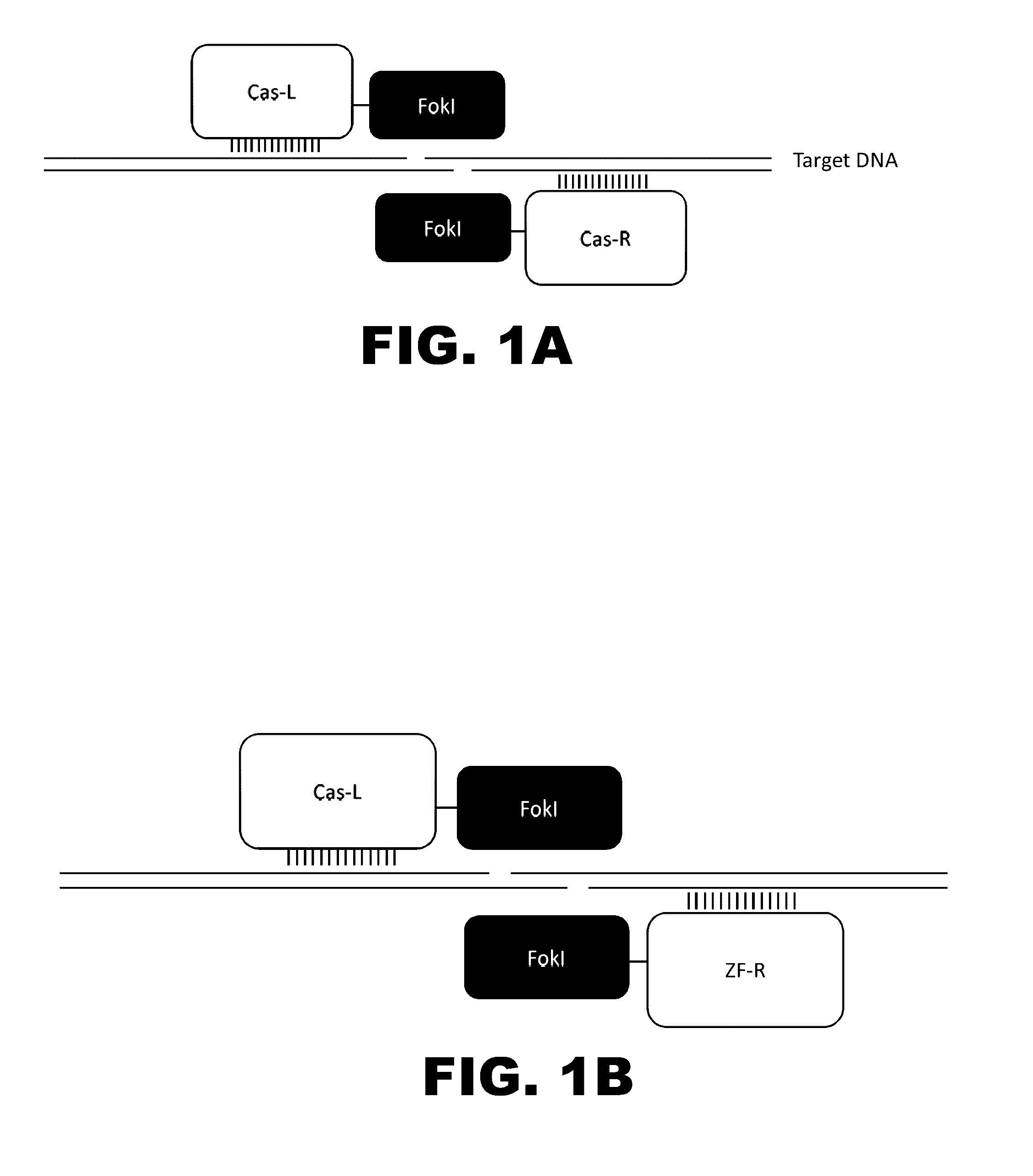
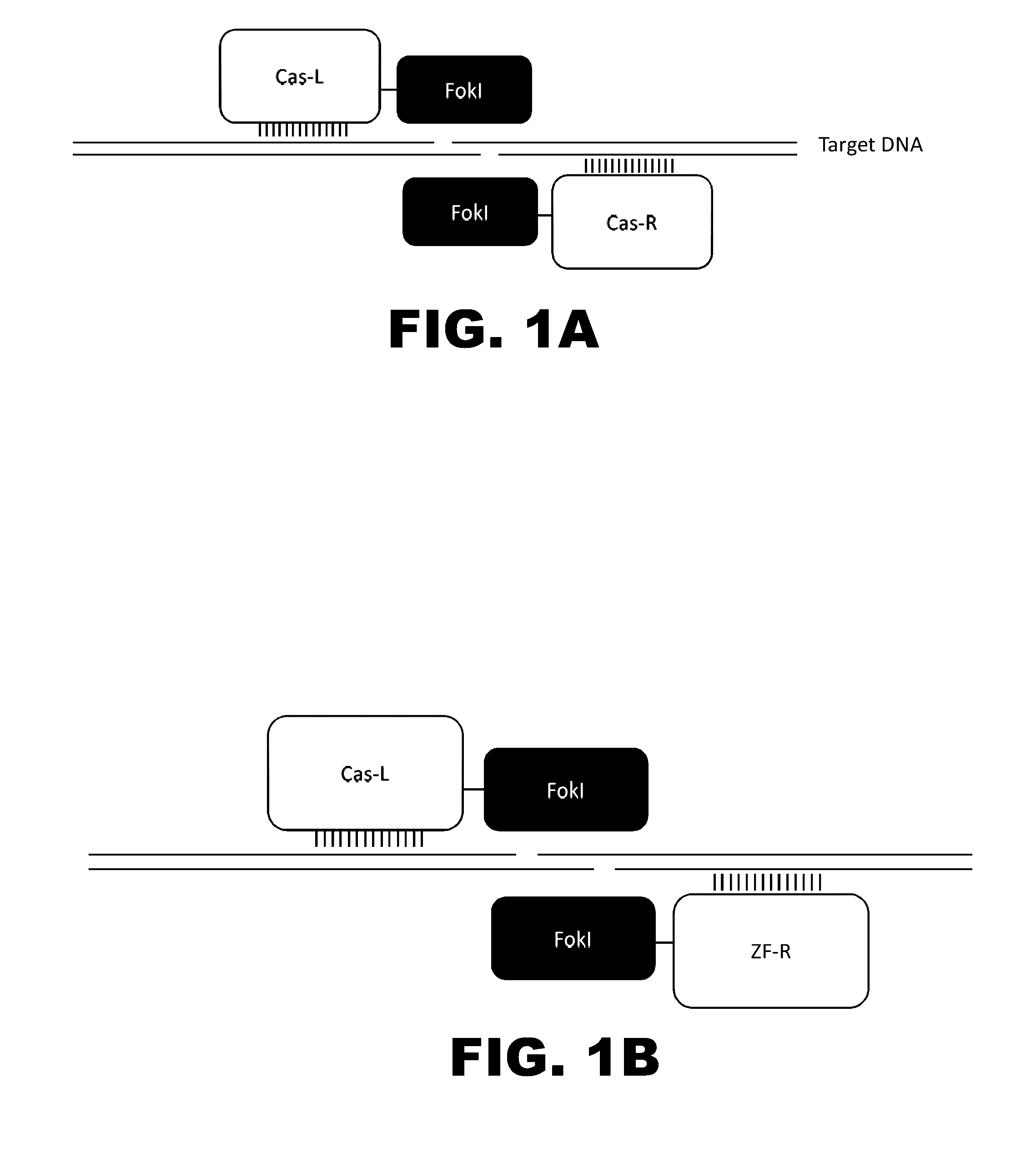
Crispr-based genome modification and regulation
Owner:SIGMA ALDRICH CO LLC
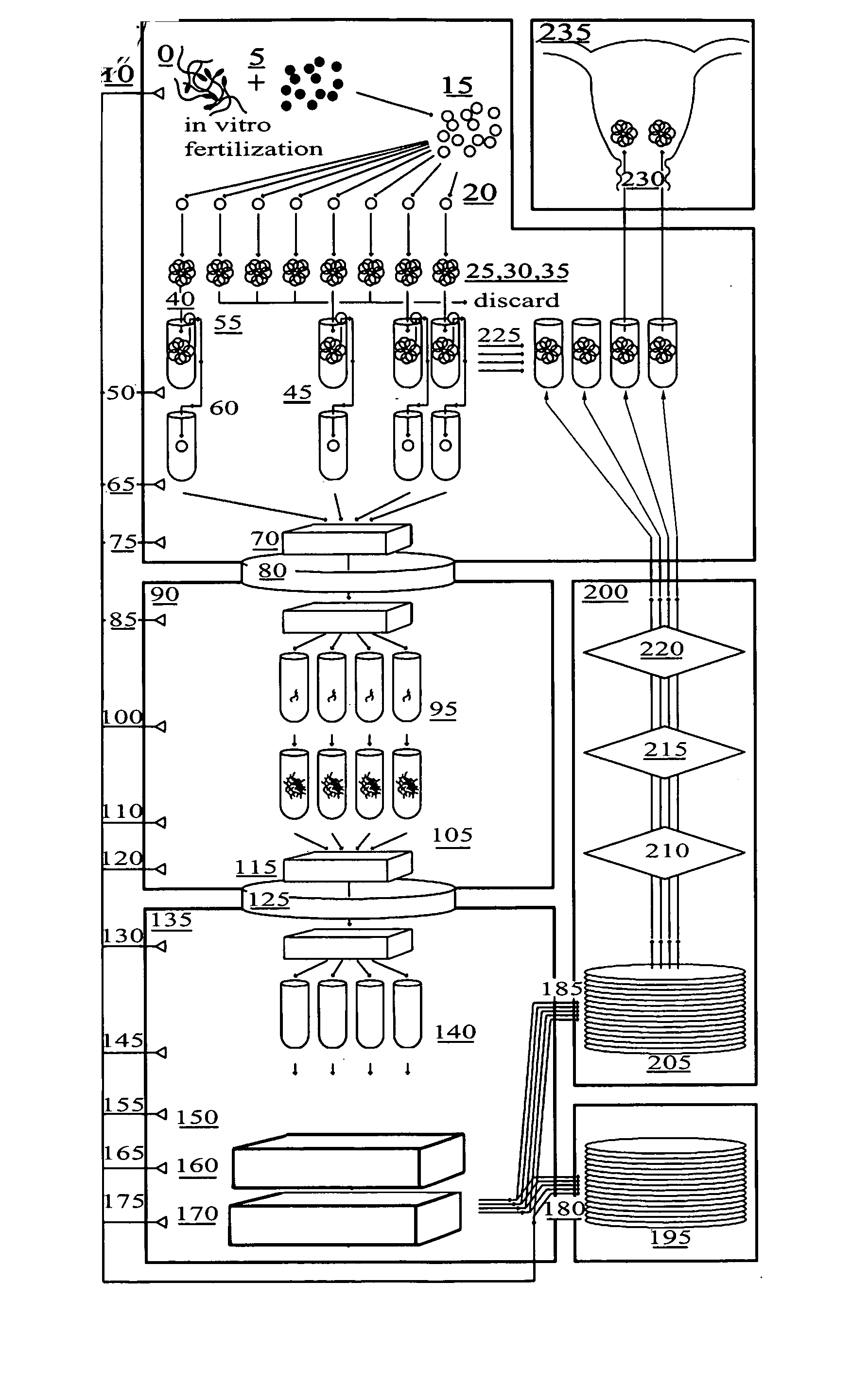
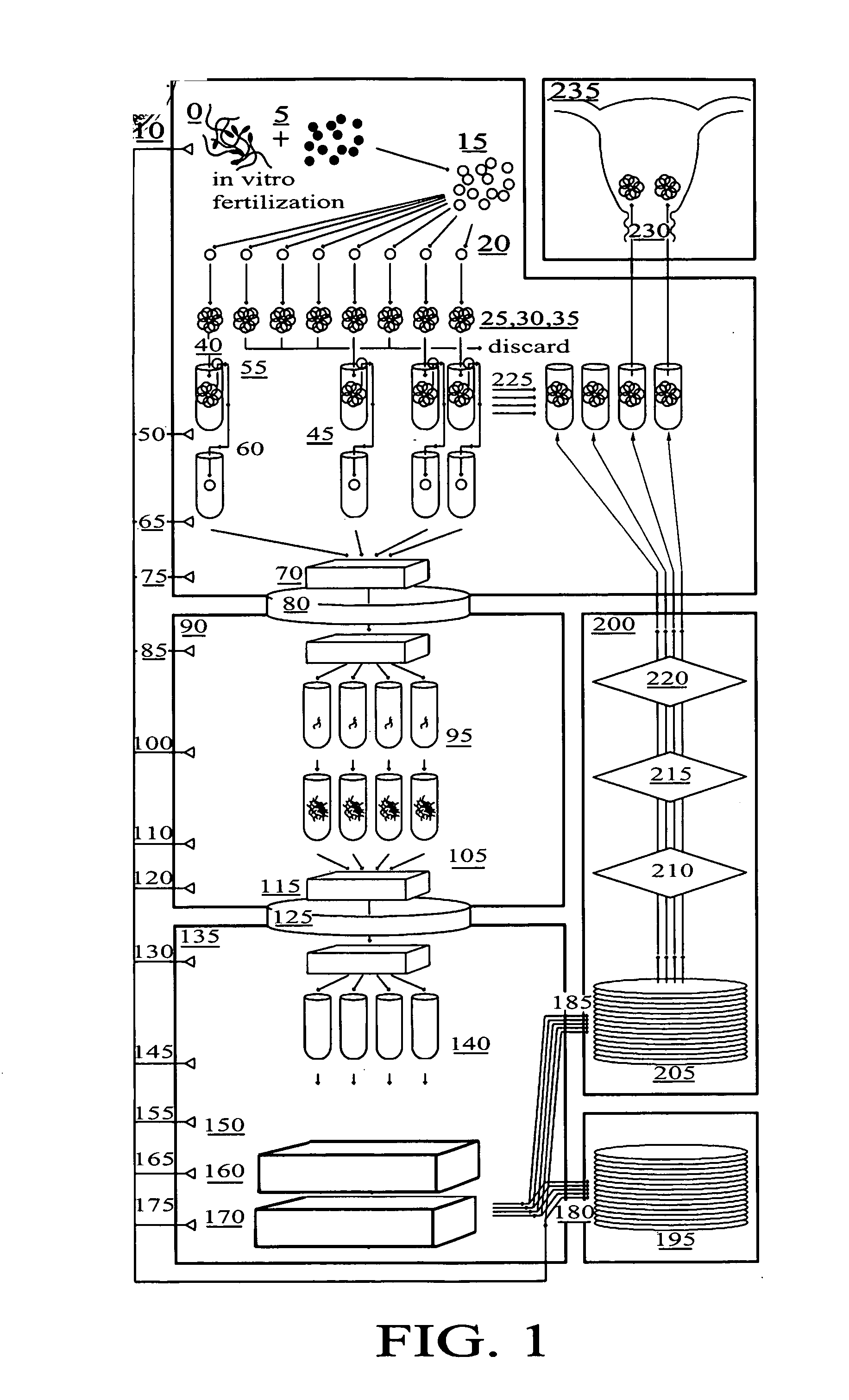
Method for genetic testing of human embryos for chromosome abnormalities, segregating genetic disorders with or without a known mutation and mitochondrial disorders following in vitro fertilization (IVF), embryo culture and embryo biopsy
InactiveUS20080085836A1Reduce significant riskImprove the level ofLibrary screeningLibrary member identificationLess invasiveContamination
We describe a method for interrogating the content and primary structure of DNA by microarray analyses and to provide comprehensive genetic screening and diagnostics prior to embryo transfer within an IVF setting. We will accomplish this by the following claims: 1) an optimized embryo grading system, 2) a less invasive embryo biopsy with reduced cellular contamination, 3) an optimized DNA amplification protocol for single cells, 4) identify aneuploidy and structural chromosome abnormalities using microarrays, 5) identifying sub-telomeric chromosome rearrangements, 6) a modified DNA fingerprinting protocol, 7) determine imprinting and epigenetic changes in developing embryos, 8) performing genome-wide scans to clarify / diagnose multi-factorial genetic disease and to determine genotype / haplotype patterns that may predict future disease, 9) determining single gene disorders with or without a known DNA mutation, 10) determining mtDNA mutations and / or the combination of mtDNA and genomic (nuclear) DNA aberrations that cause genetic disease.
Owner:KEARNS WILLIAM G +1
Crispr-based genome modification and regulation
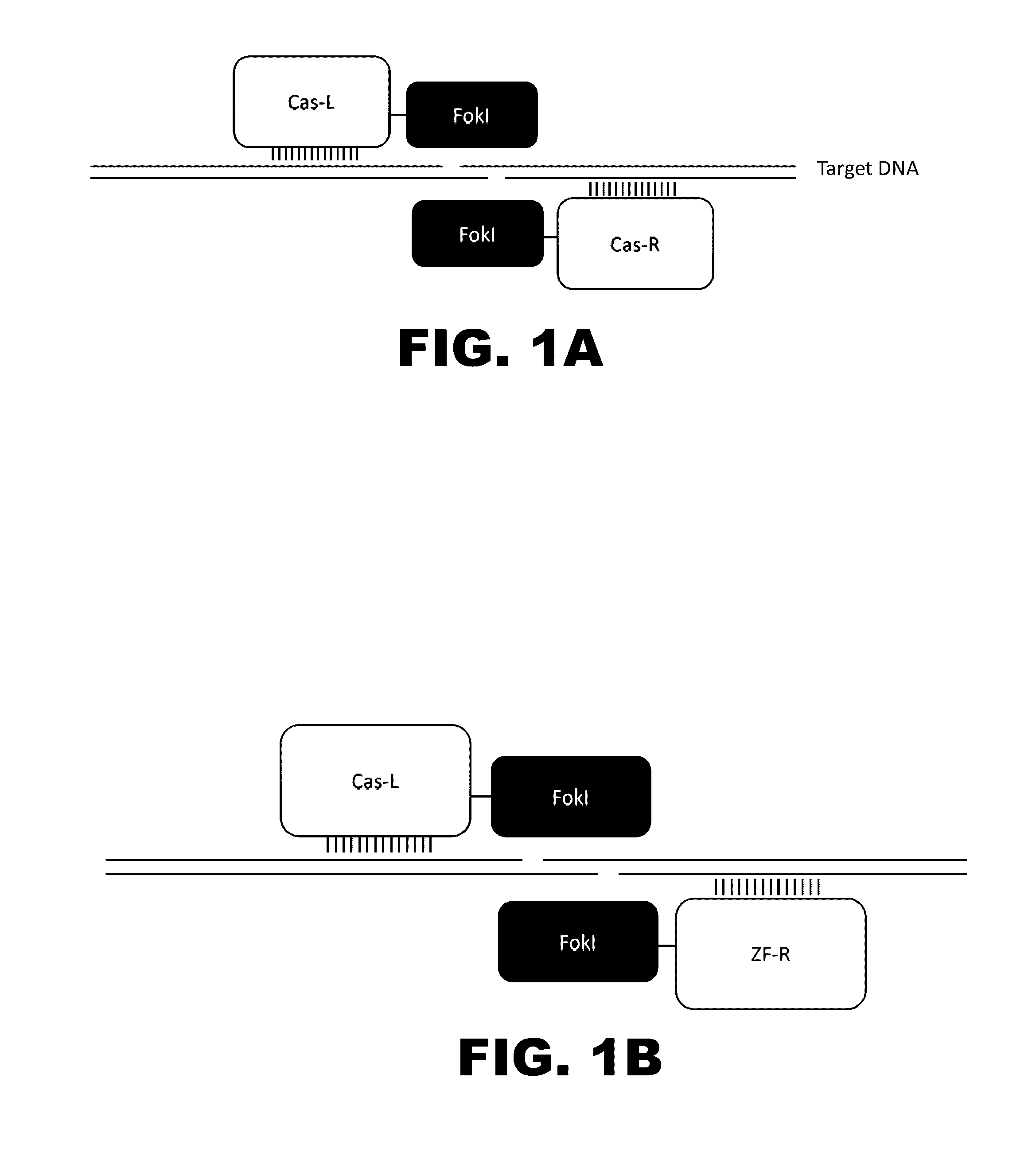
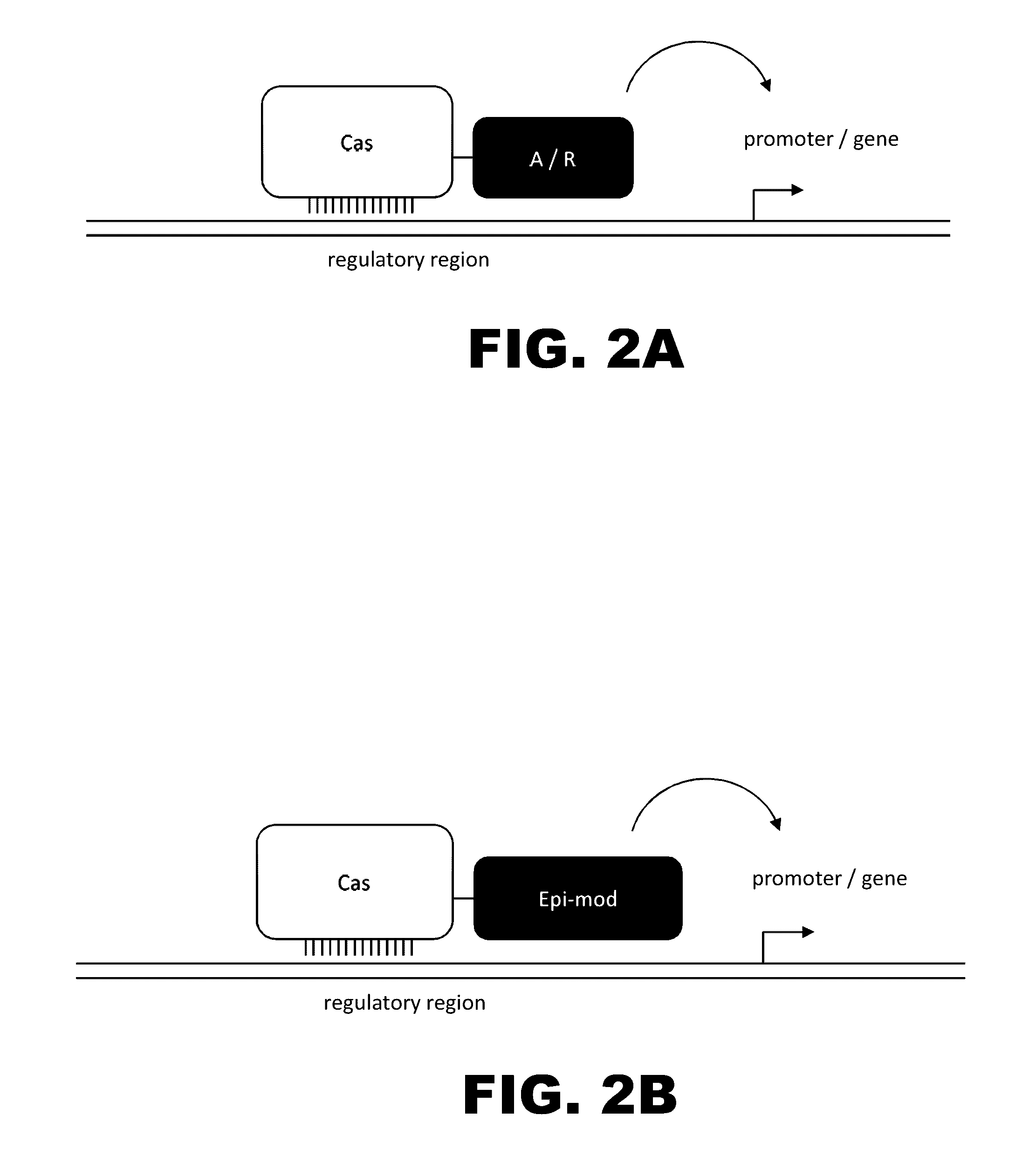
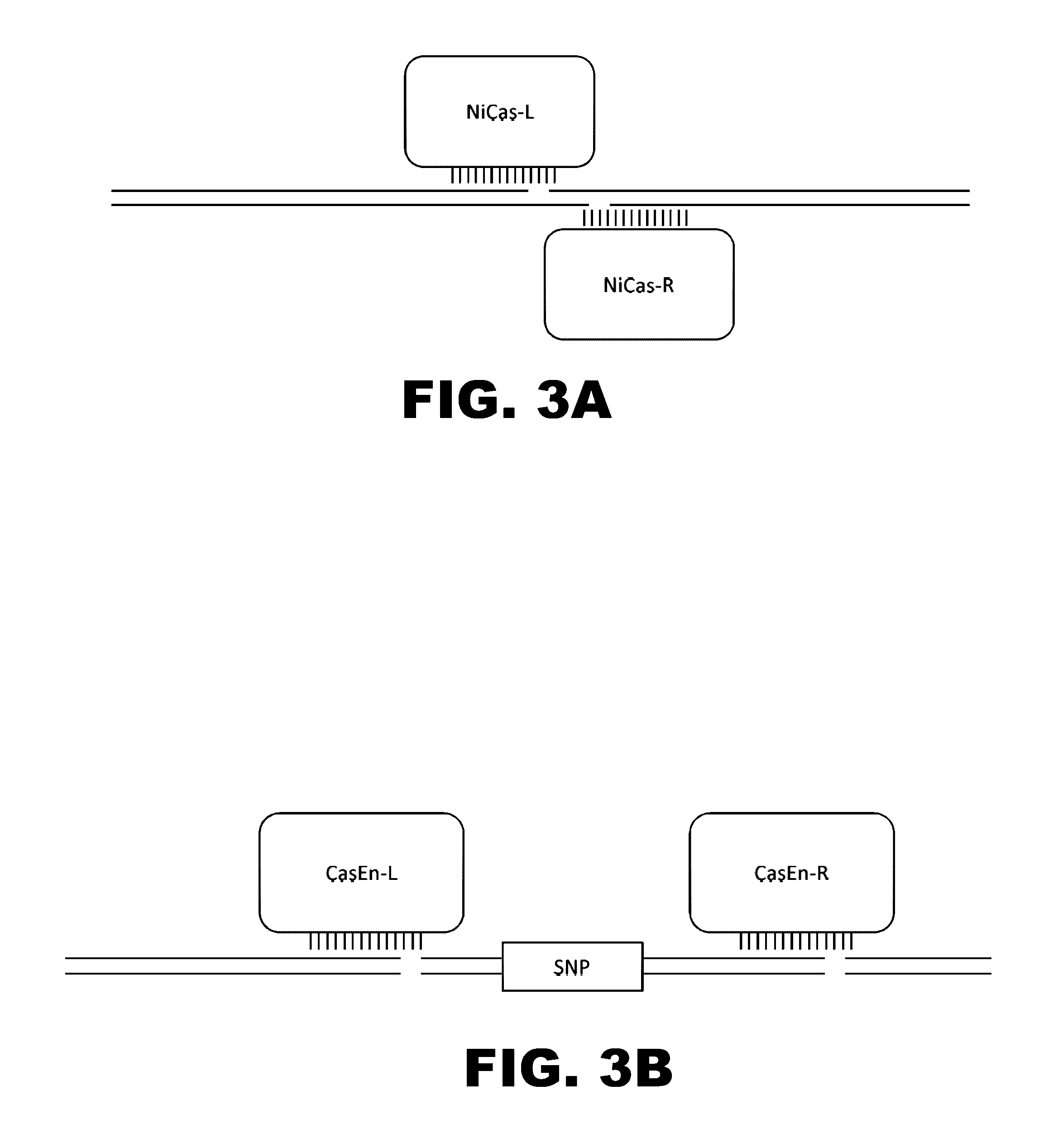
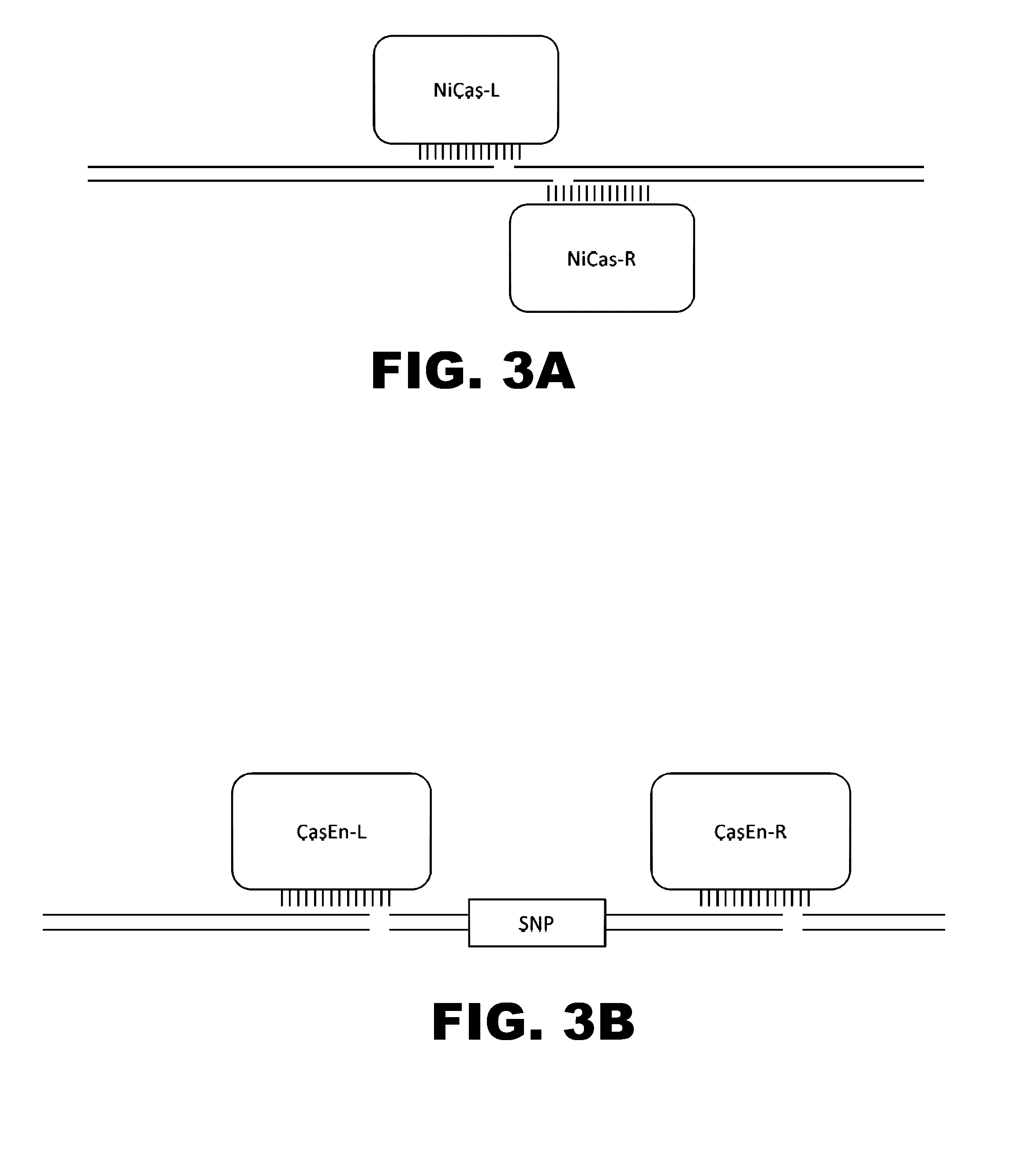
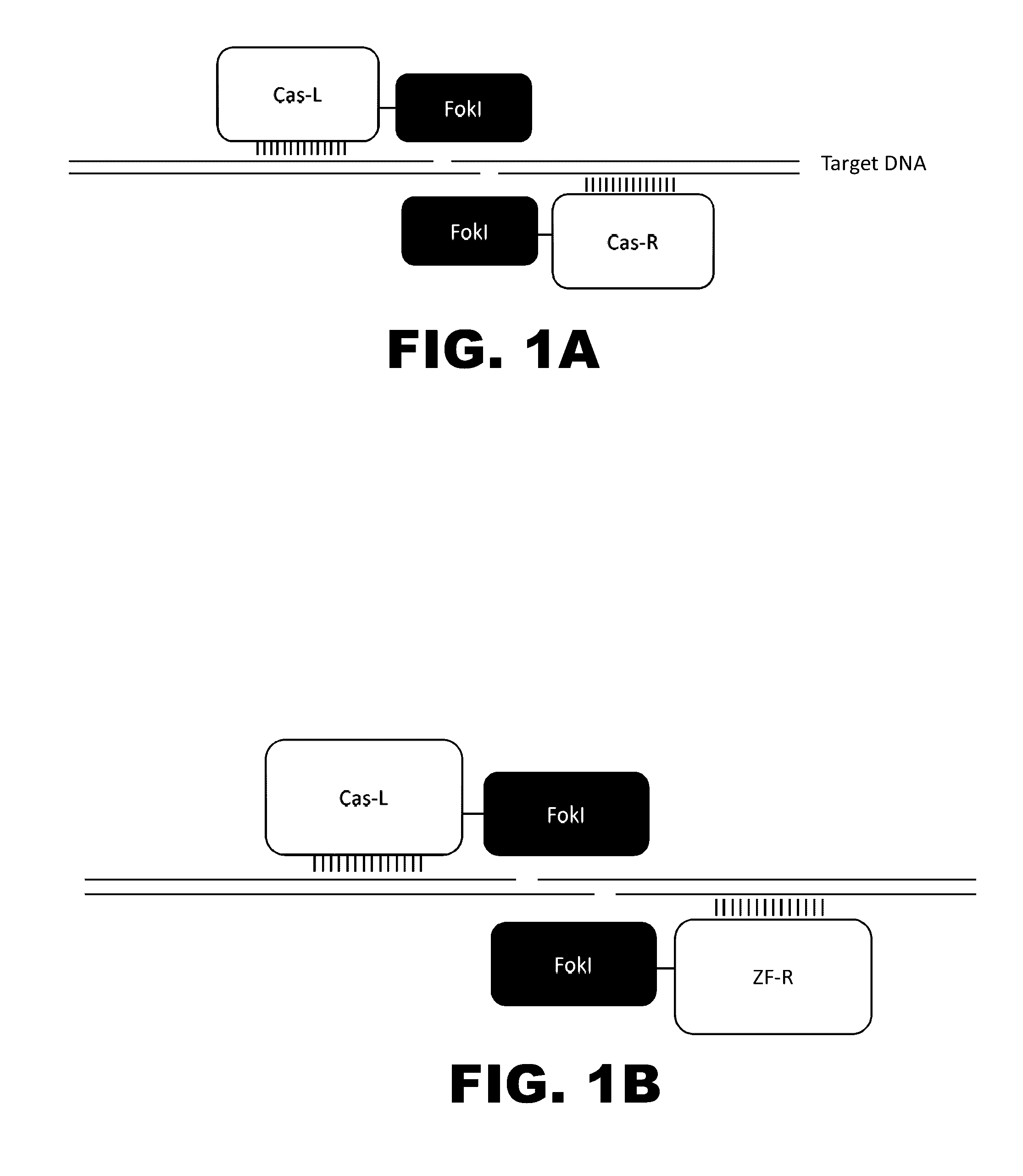
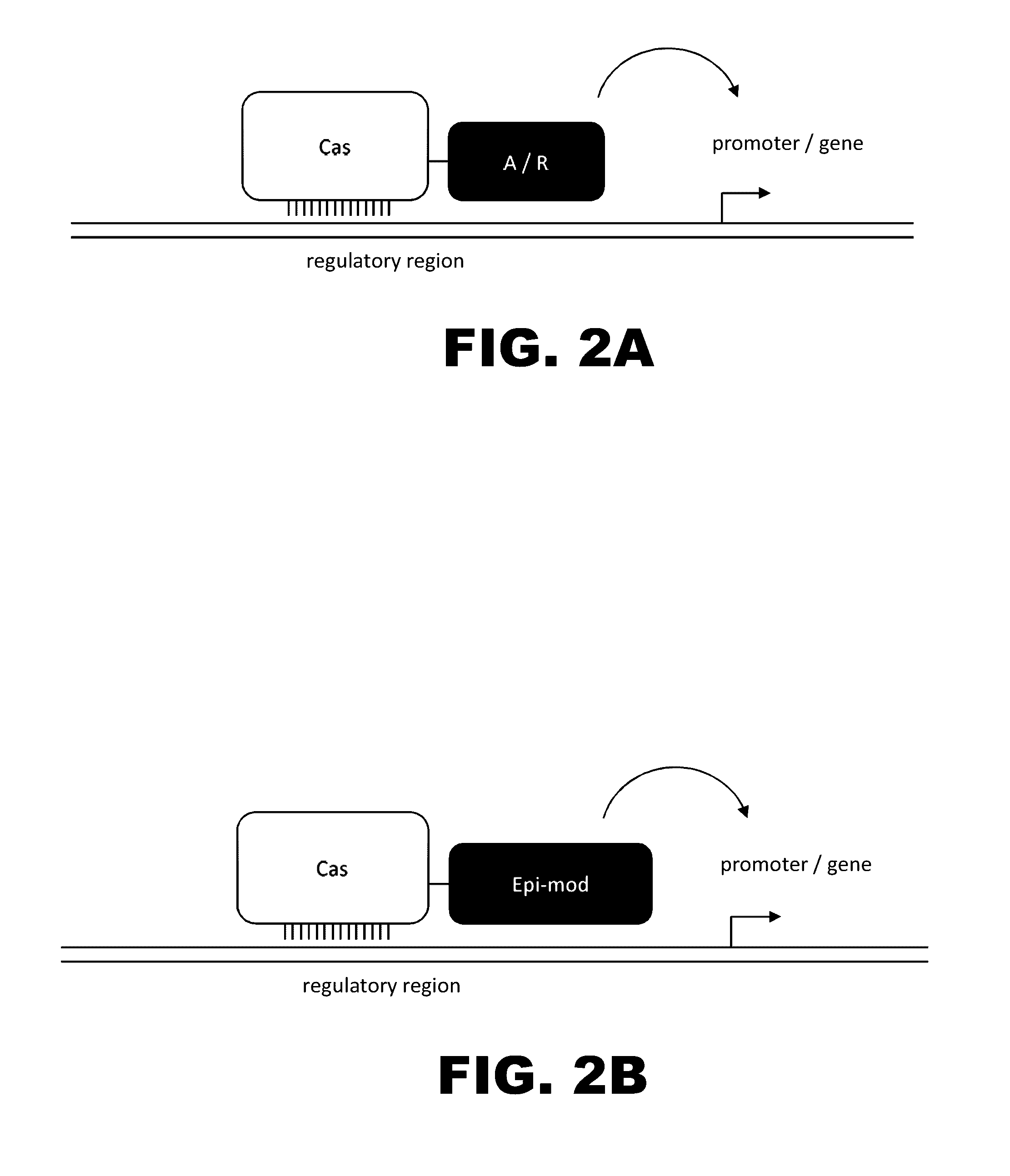
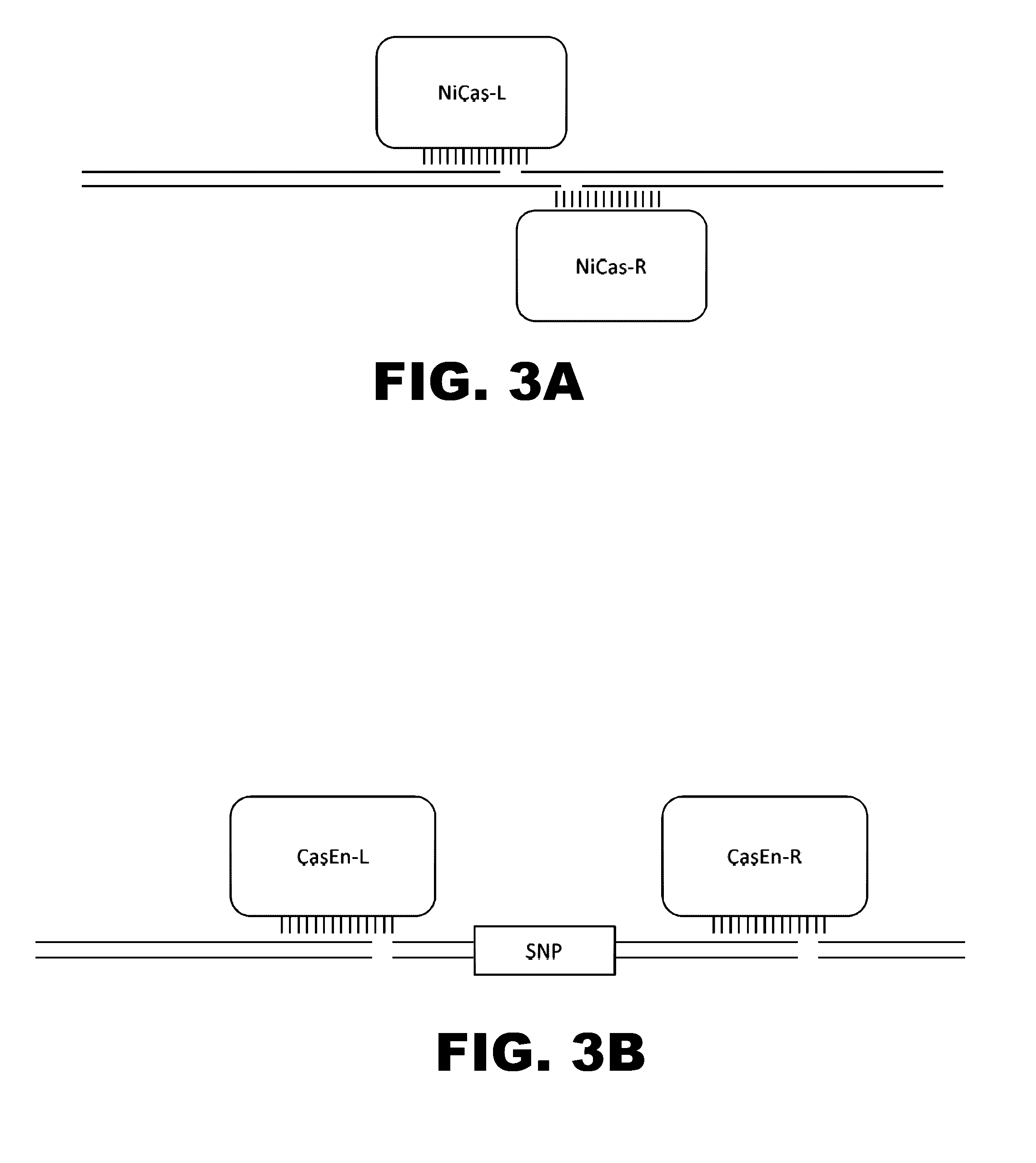
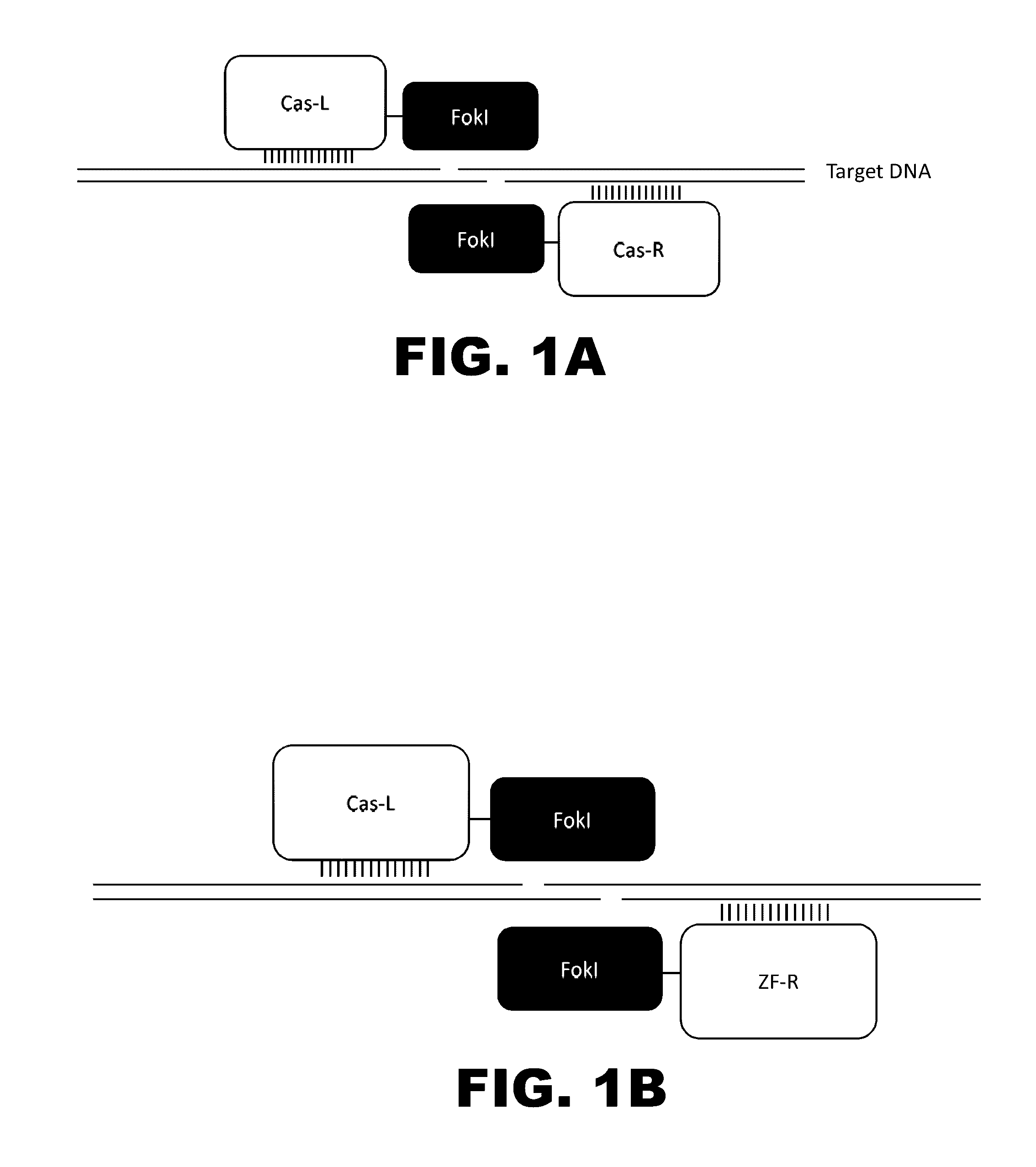
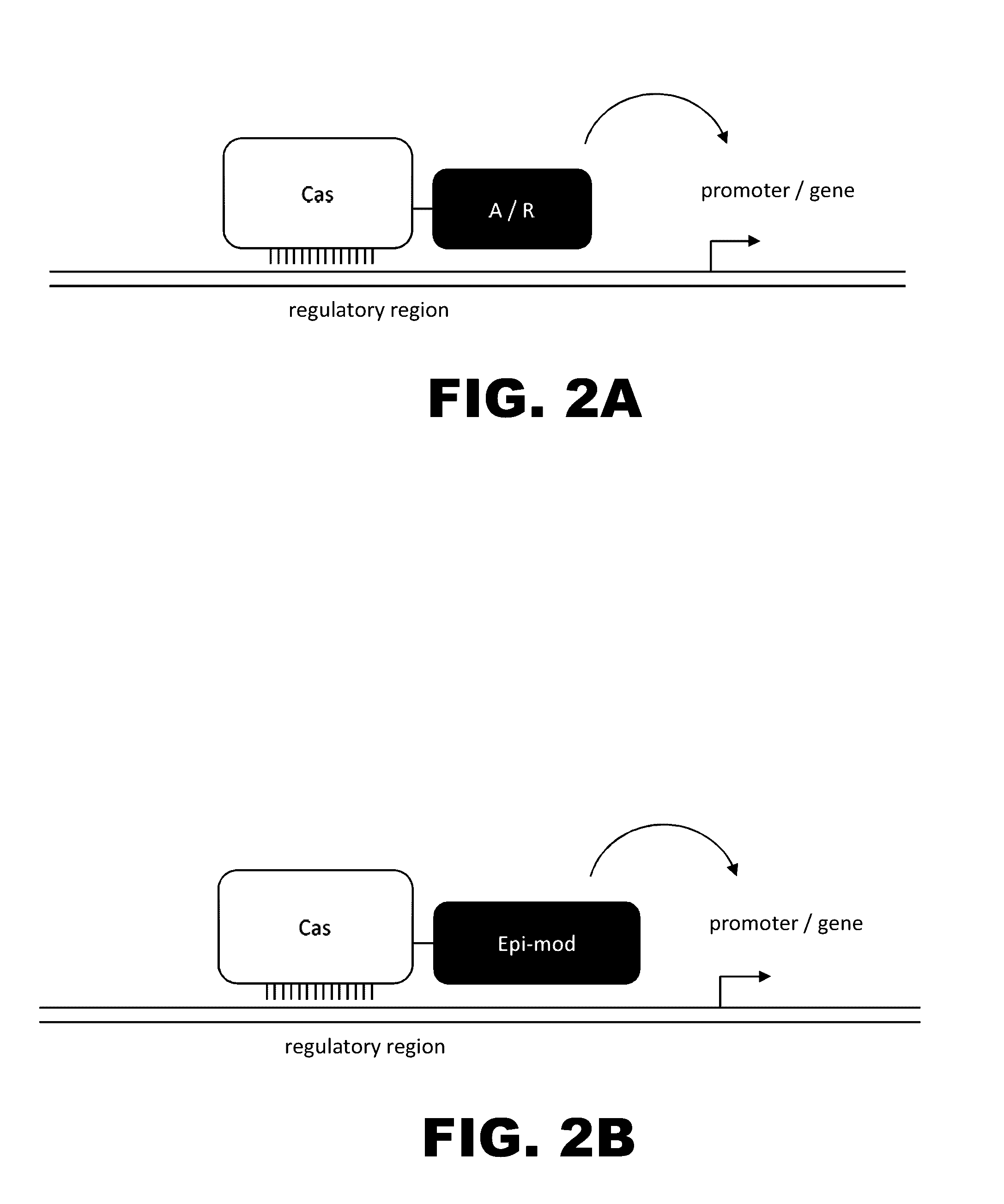
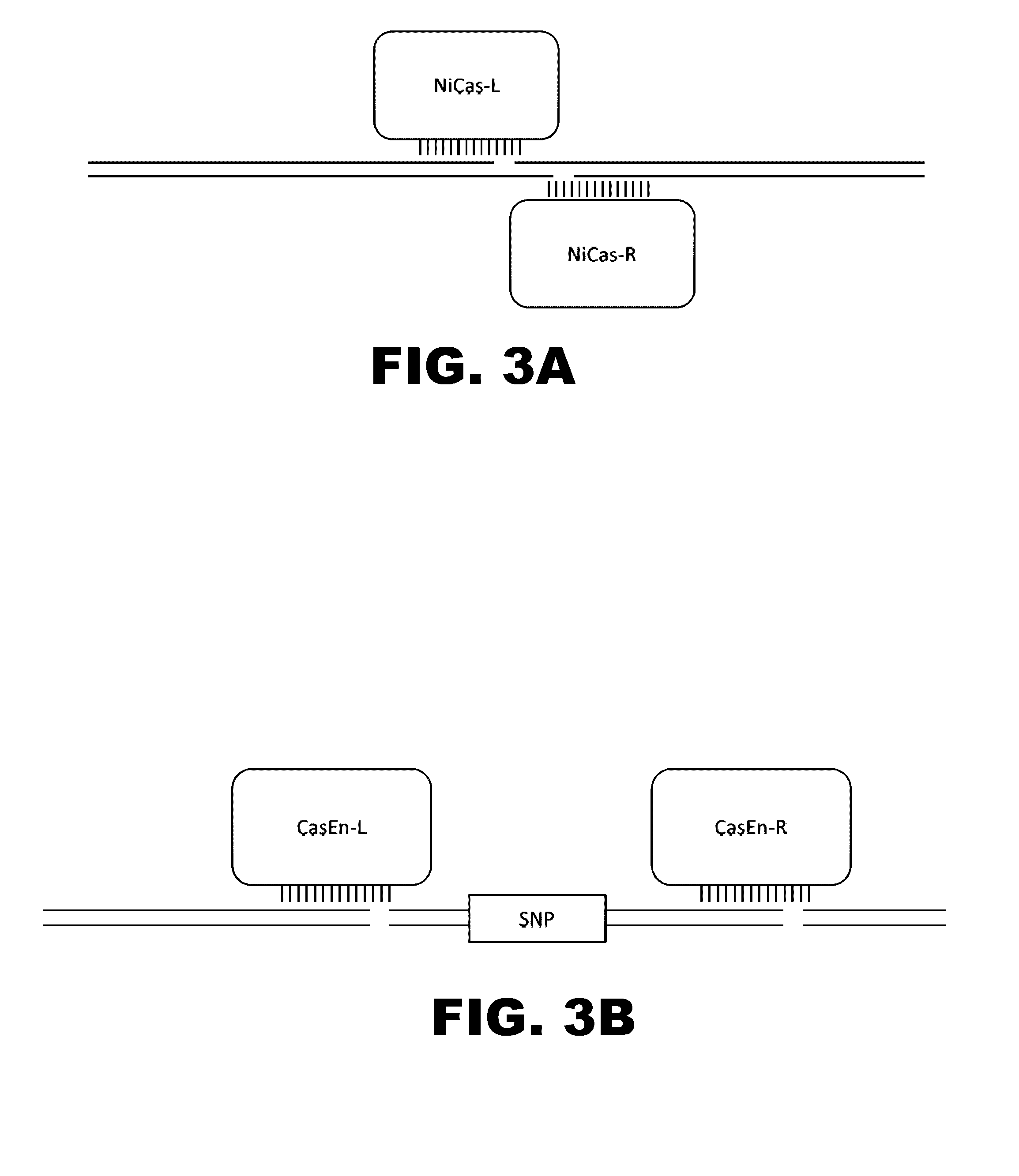
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
Diagnosis of diseases associated with the immune system by determining cytosine methylation
InactiveUS20030143606A1Bioreactor/fermenter combinationsBiological substance pretreatmentsCytosineOligomer
The present invention relates to chemically modified genomic sequences of genes associated with the immune system, to oligonucleotides and / or PNA-oligomers directed against the sequence, for the detection of the methylation status of genes, associated with the immune system as well as to a method for ascertaining genetic and / or epigentic parametres of genes, associated with the immune system.
Owner:EPIGENOMICS AG
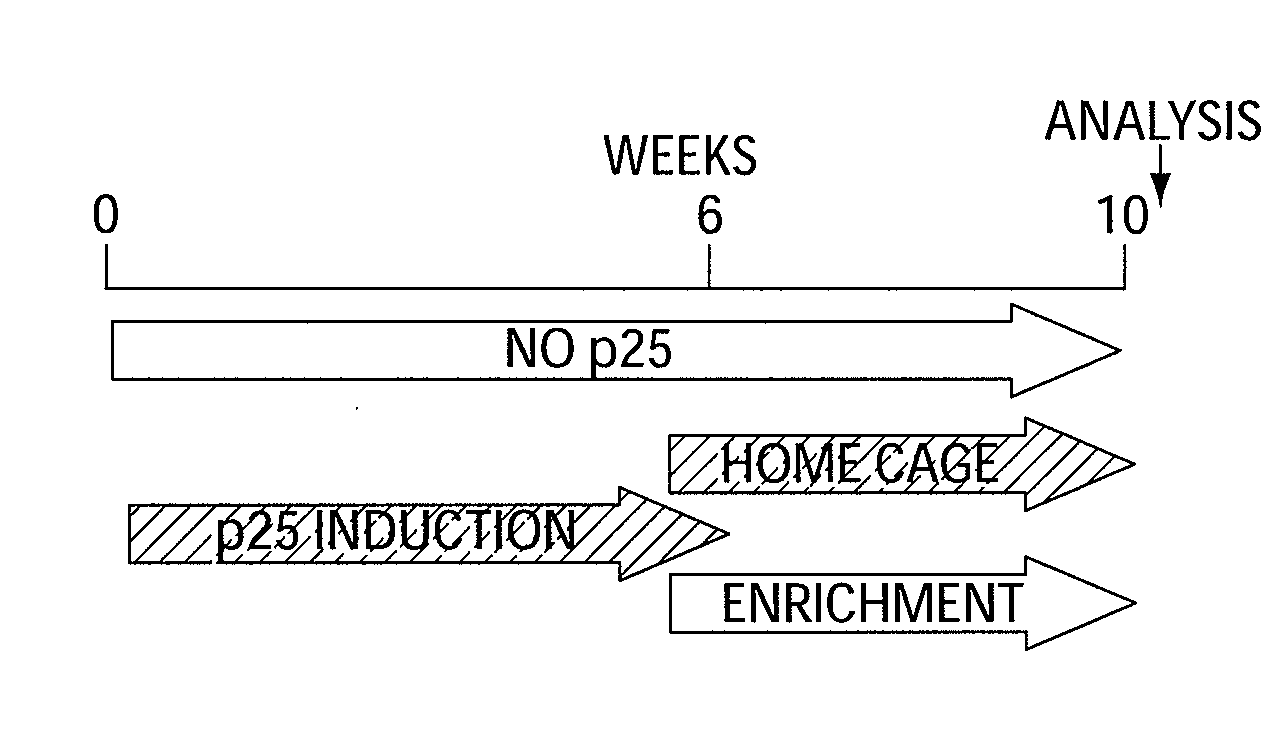
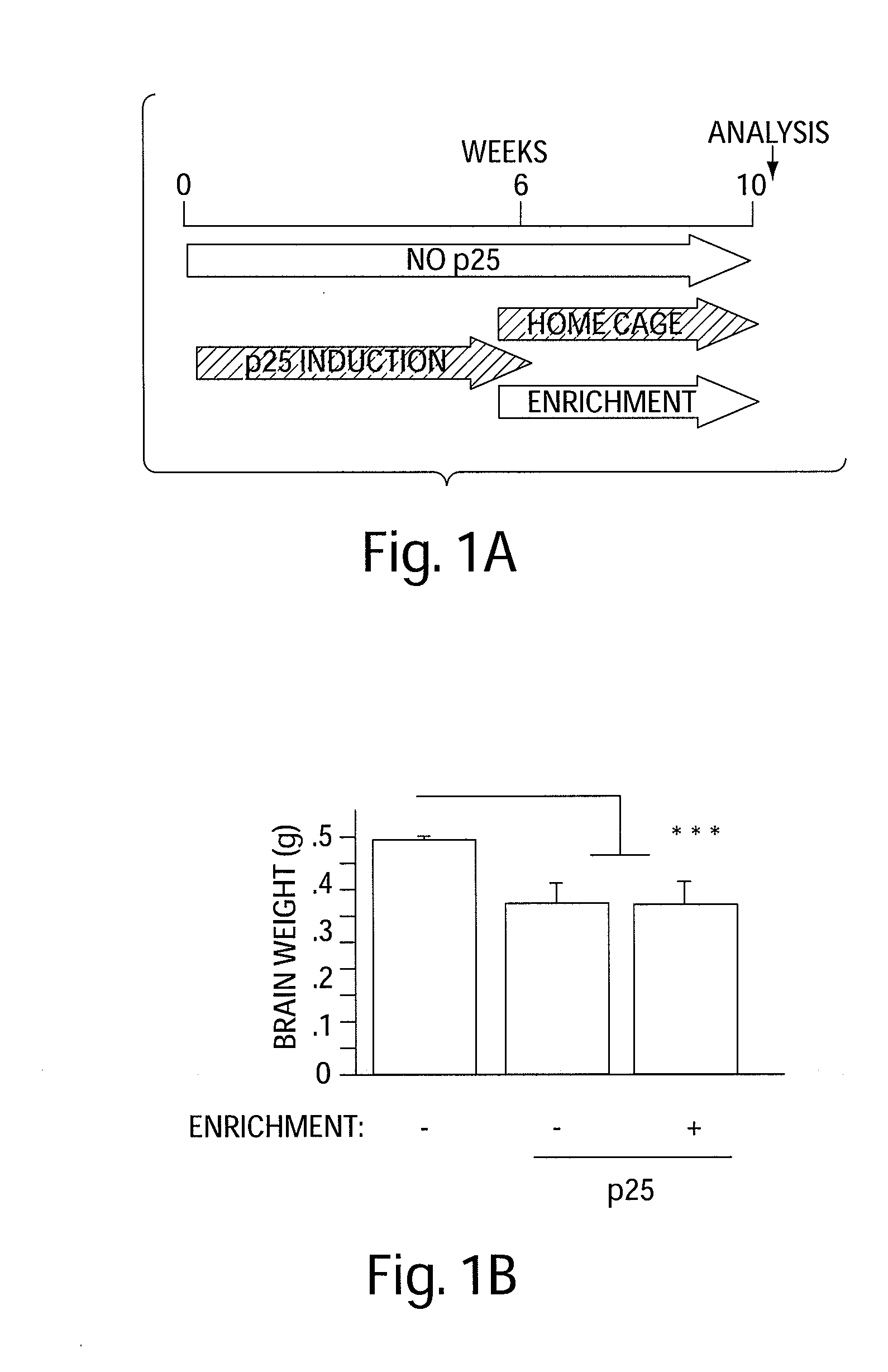
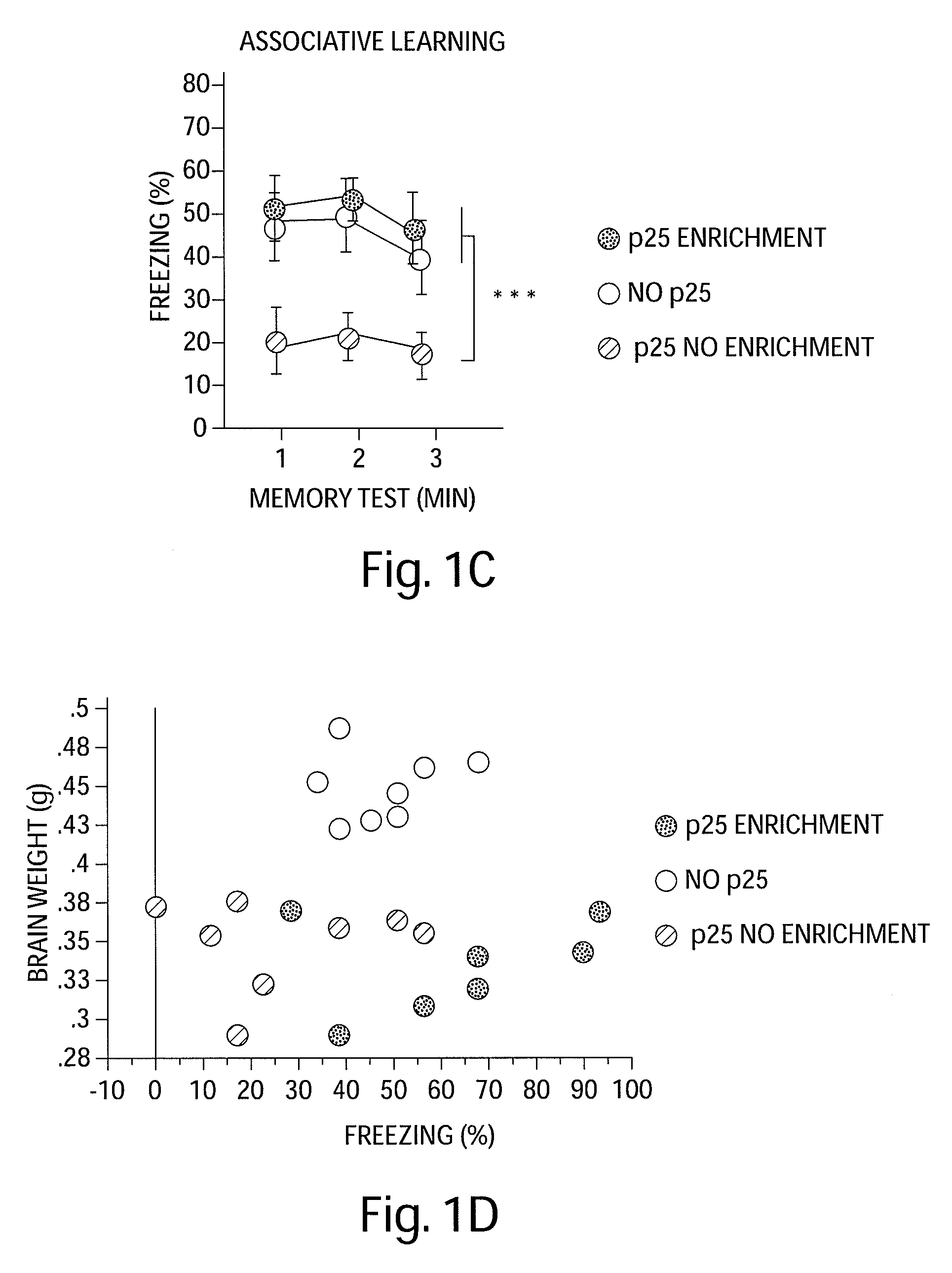
Epigenetic mechanisms re-establish access to long-term memory after neuronal loss
ActiveUS20080300205A1Increase the number ofIncrease in histone acetylationBiocideNervous disorderEpigenetic ProfileAcetylation
The invention relates to methods and products for enhancing and improving recovery of lost memories. In particular the methods are accomplished through the increase of histone acetylation.
Owner:THE GENERAL HOSPITAL CORP +2
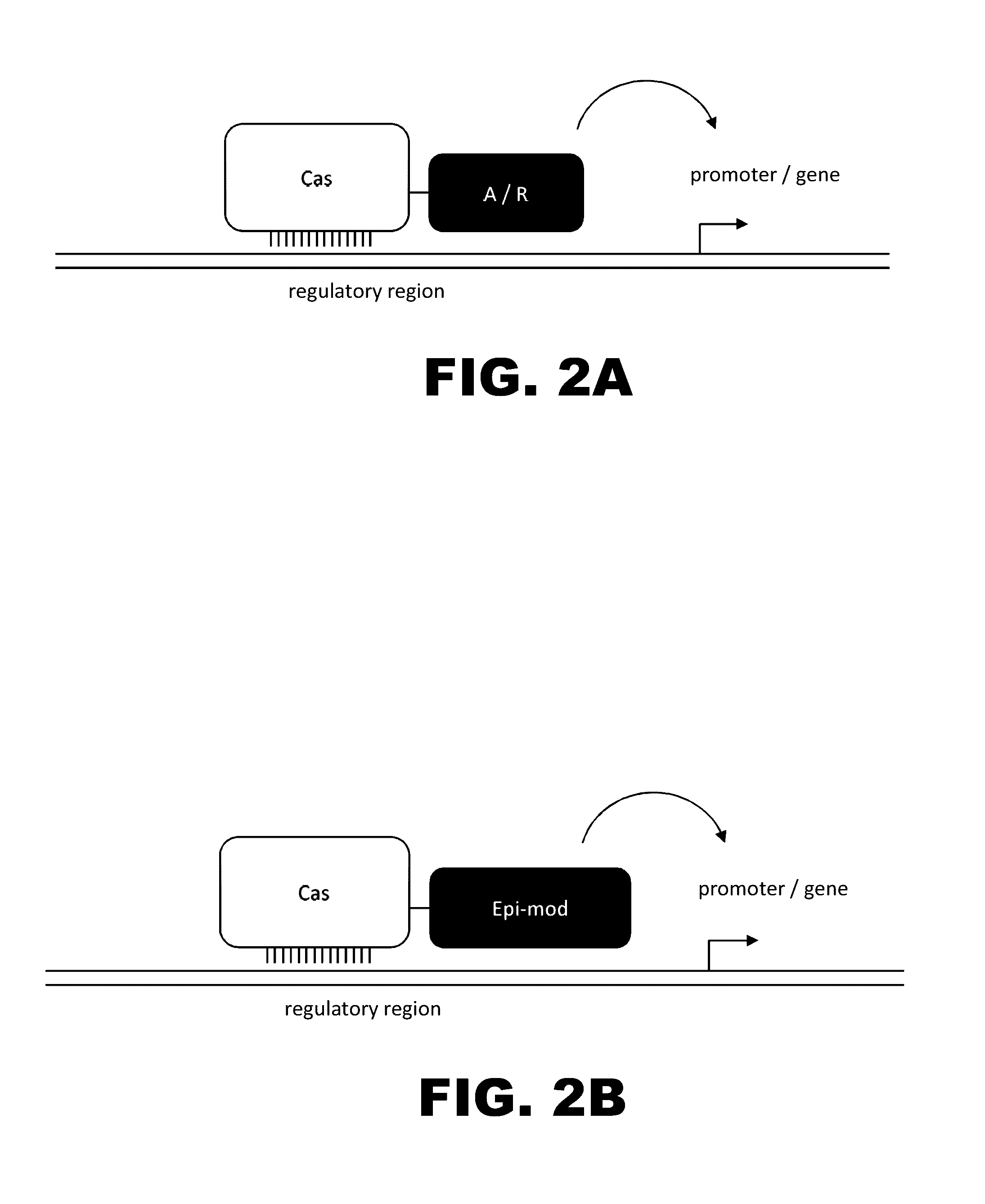
Crispr-based genome modification and regulation
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
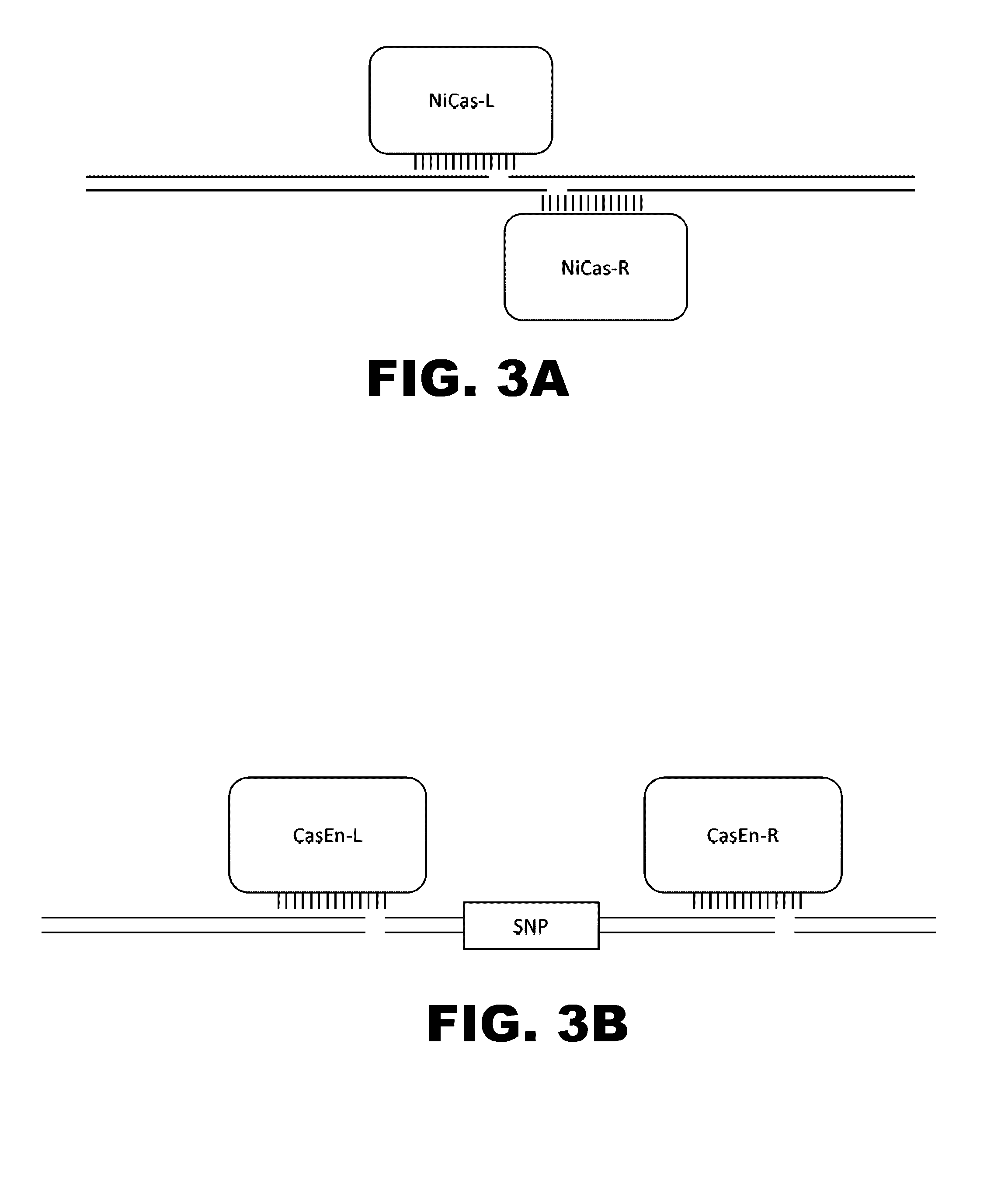
Crispr-based genome modification and regulation
Owner:SIGMA ALDRICH CO LLC
Crispr-based genome modification and regulation
The present invention provides RNA-guided endonucleases, which are engineered for expression in eukaryotic cells or embryos, and methods of using the RNA-guided endonuclease for targeted genome modification in eukaryotic cells or embryos. Also provided are fusion proteins, wherein each fusion protein comprises a CRISPR / Cas-like protein or fragment thereof and an effector domain. The effector domain can be a cleavage domain, an epigenetic modification domain, a transcriptional activation domain, or a transcriptional repressor domain. Also provided are methods for using the fusion proteins to modify a chromosomal sequence or regulate expression of a chromosomal sequence.
Owner:SIGMA ALDRICH CO LLC
Methods and compositions for differentiating tissues for cell types using epigenetic markers
InactiveUS20060183128A1Accurate and efficientMicrobiological testing/measurementFermentationGenomic DNAEpigenetic Profile
The present invention provides, inter alia, a method for generating a genome-wide epigenomic map, comprising a correlation between methylation variable CpG positions (MVP) and genomic DNA sample types. MVP are those CpG positions that show a variable quantitative level of methylation between sample types. Particular genomic regions of interest (ROI) provide preferred marker sequences that comprise multiple, and preferably proximate MVP, and that have novel utility for distinguishing sample types. The epigenic maps have broad utility, for example, in identifying sample types, or for distinguishing between and among sample types. In a preferred embodiment the epigenomic map is based on methylation variable regions (MVP) within the major histocompatibility complex (MHC), and has utility, for example, in identifying the cell or tissue source of a genomic DNA sample, or for distinguishing one or more particular cell or tissue types among other cell or tissue types. Analysis of epigenetic characteristics of one, or of a set of nucleic acid sequences, in the context of an inventive epigenomic map, allows for the determination of an origin of the nucleic acids.
Owner:EPIGENOMICS AG
Diagnosis of diseases associated with metastasis
The present invention relates to the chemically modified genomic sequences of genes associated with metastasis, to oligonucleotides and / or PNA-oligomers for detecting the cytosine methylation state of genes associated with metastasis which are directed against the sequence, as well as to a method for ascertaining genetic and / or epigenetic parameters of genes associated with metastasis.
Owner:EPIGENOMICS AG
Methods and materials for detecting colorectal cancer and adenoma
InactiveUS20130012410A1High detectionHigh level of sensitivityMicrobiological testing/measurementLibrary screeningFOXE1Mammal
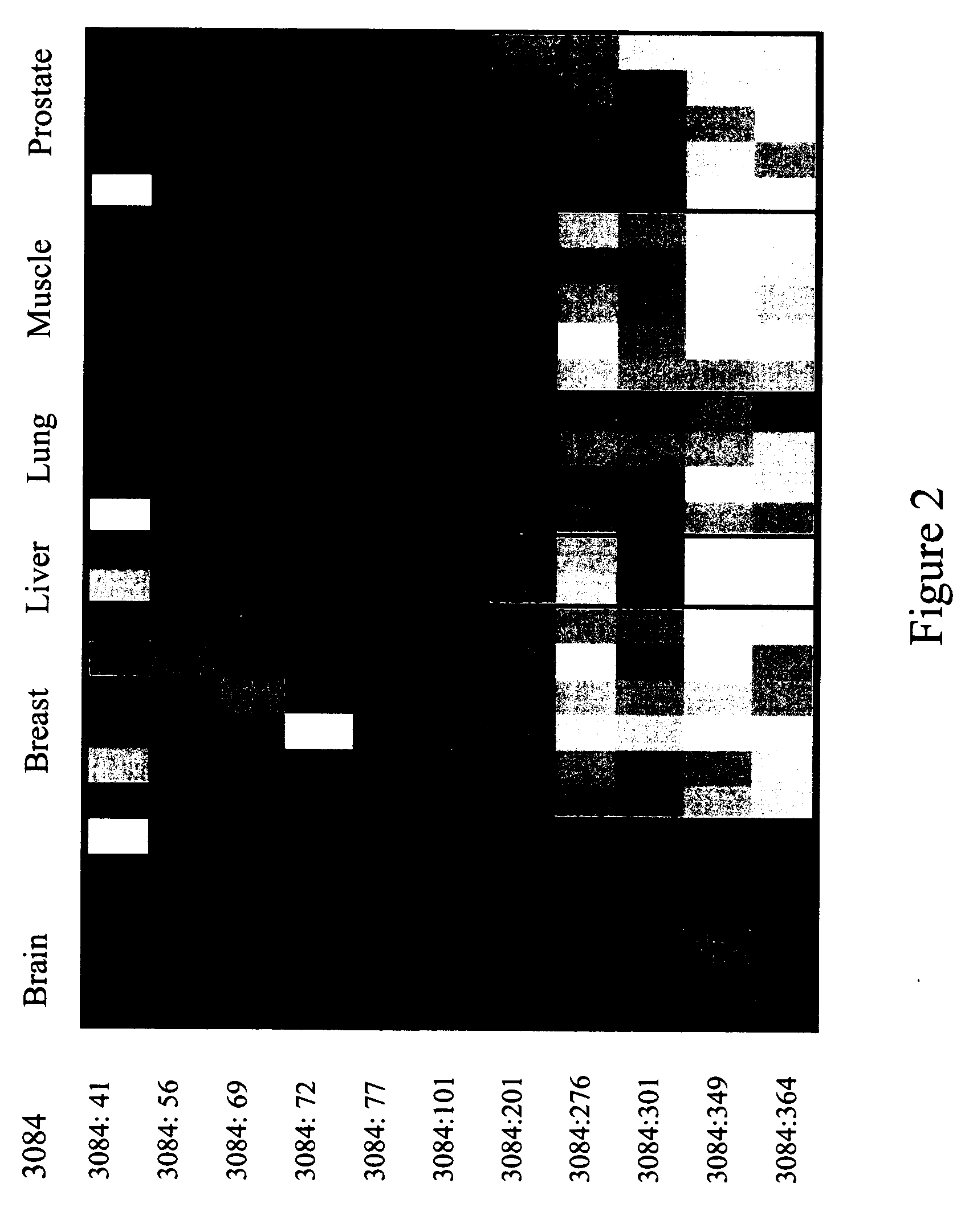
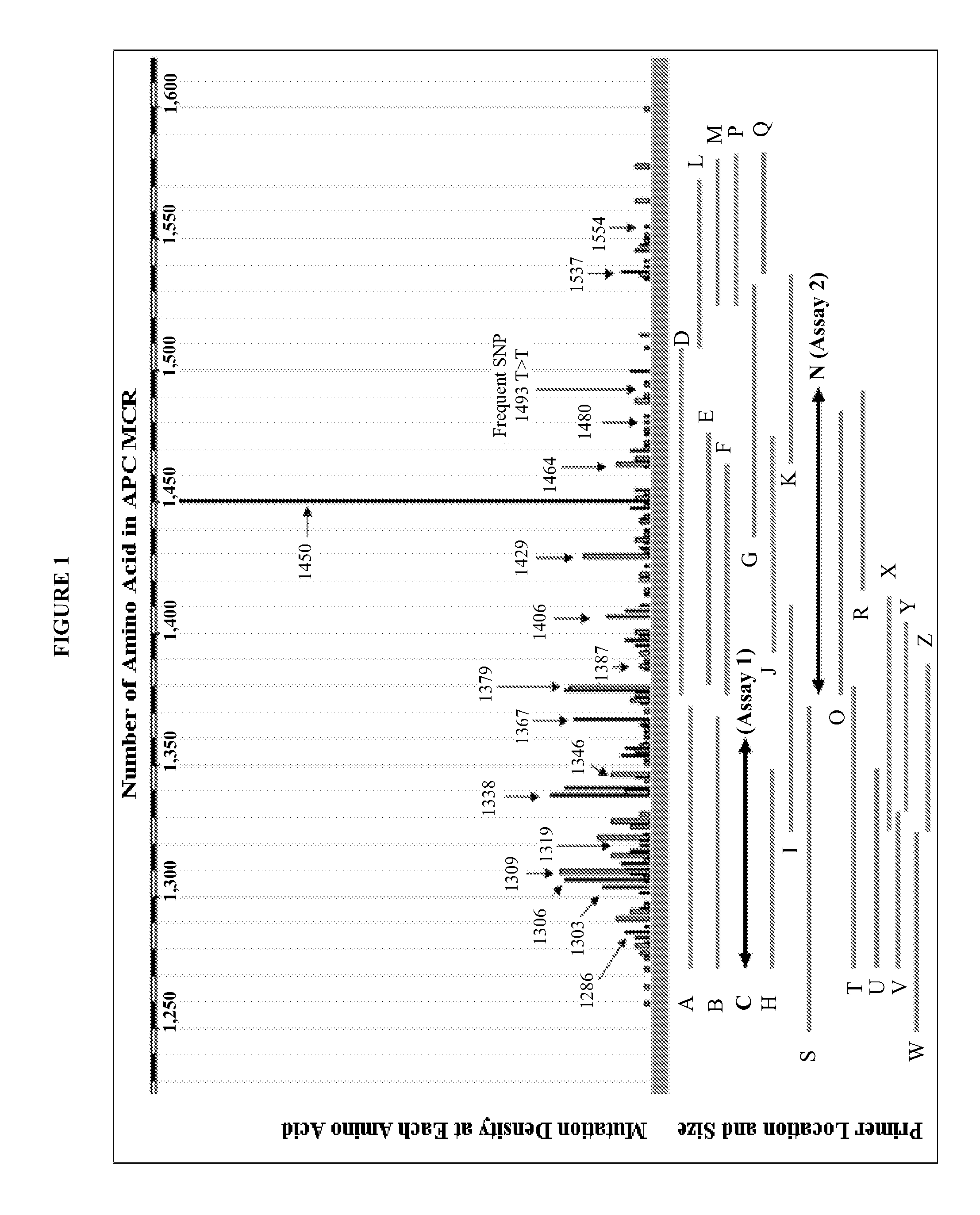
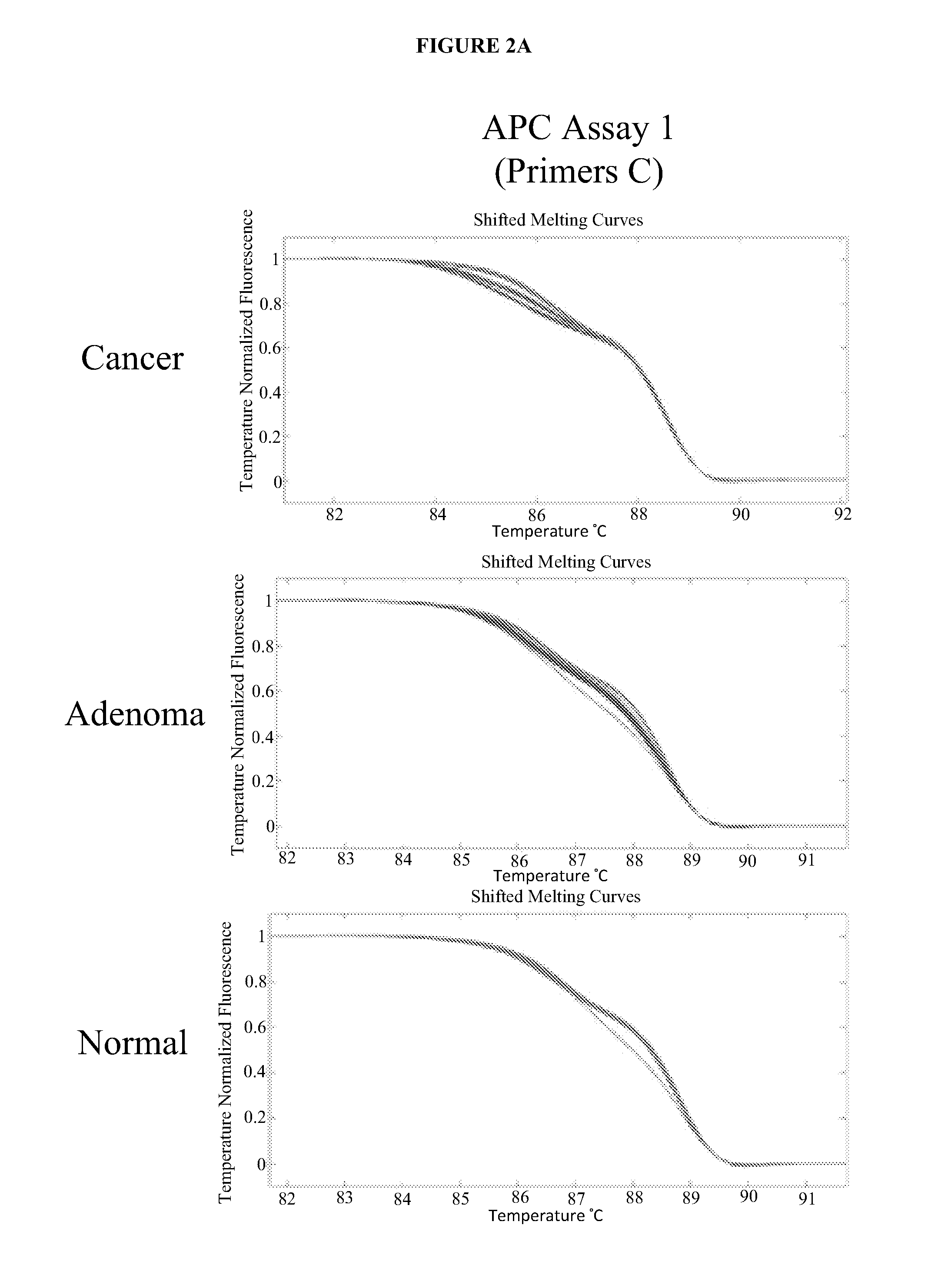
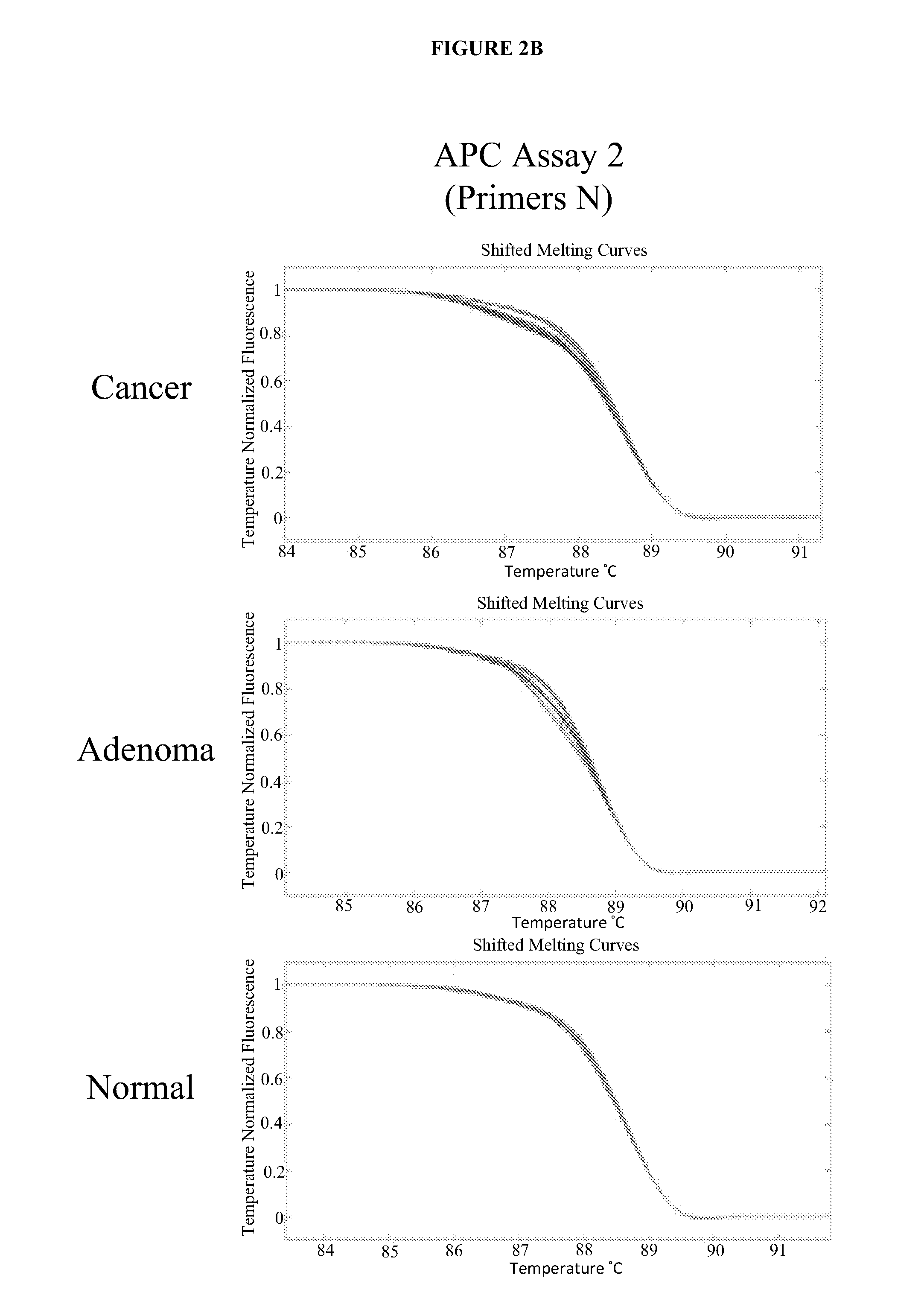
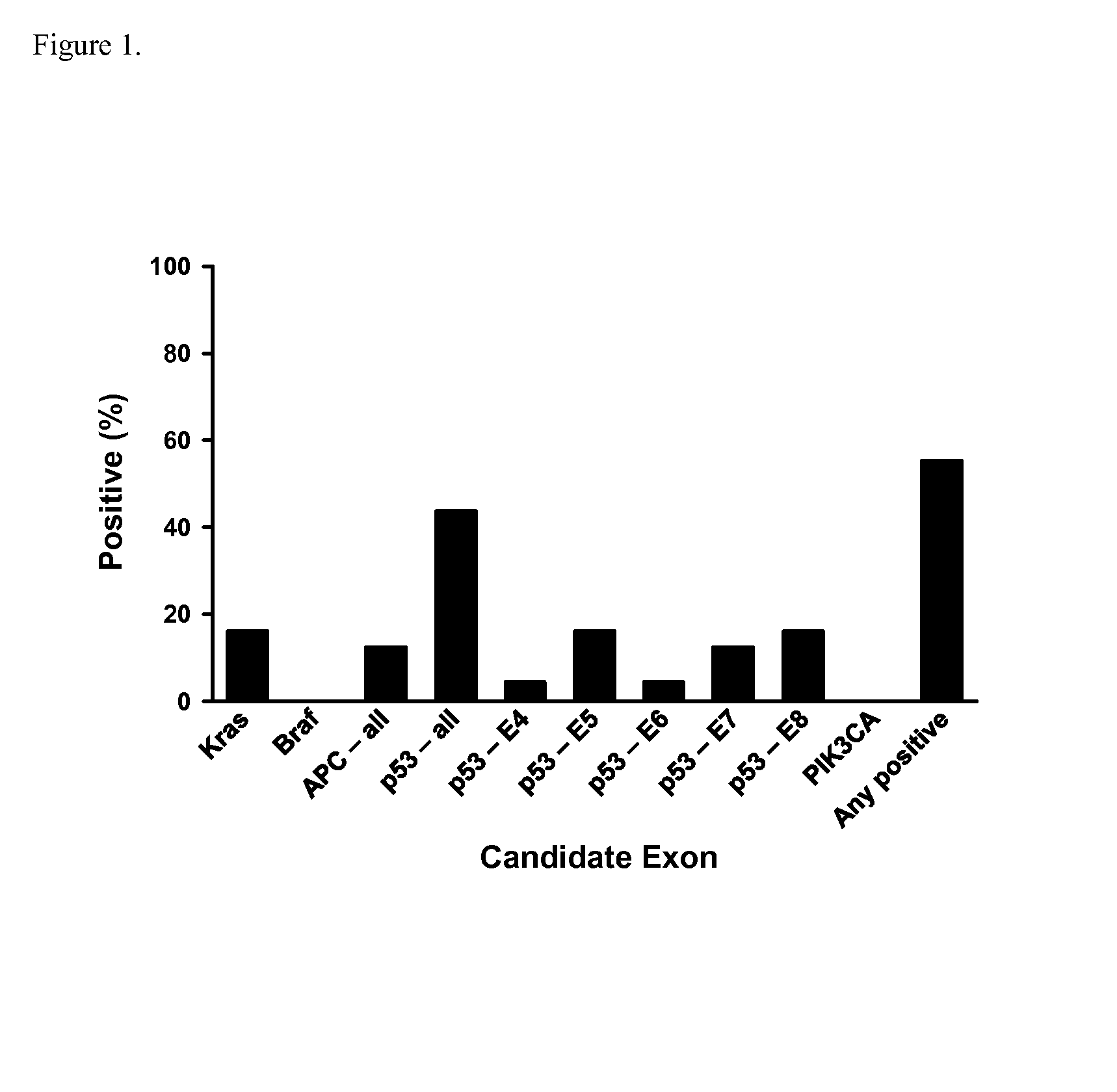
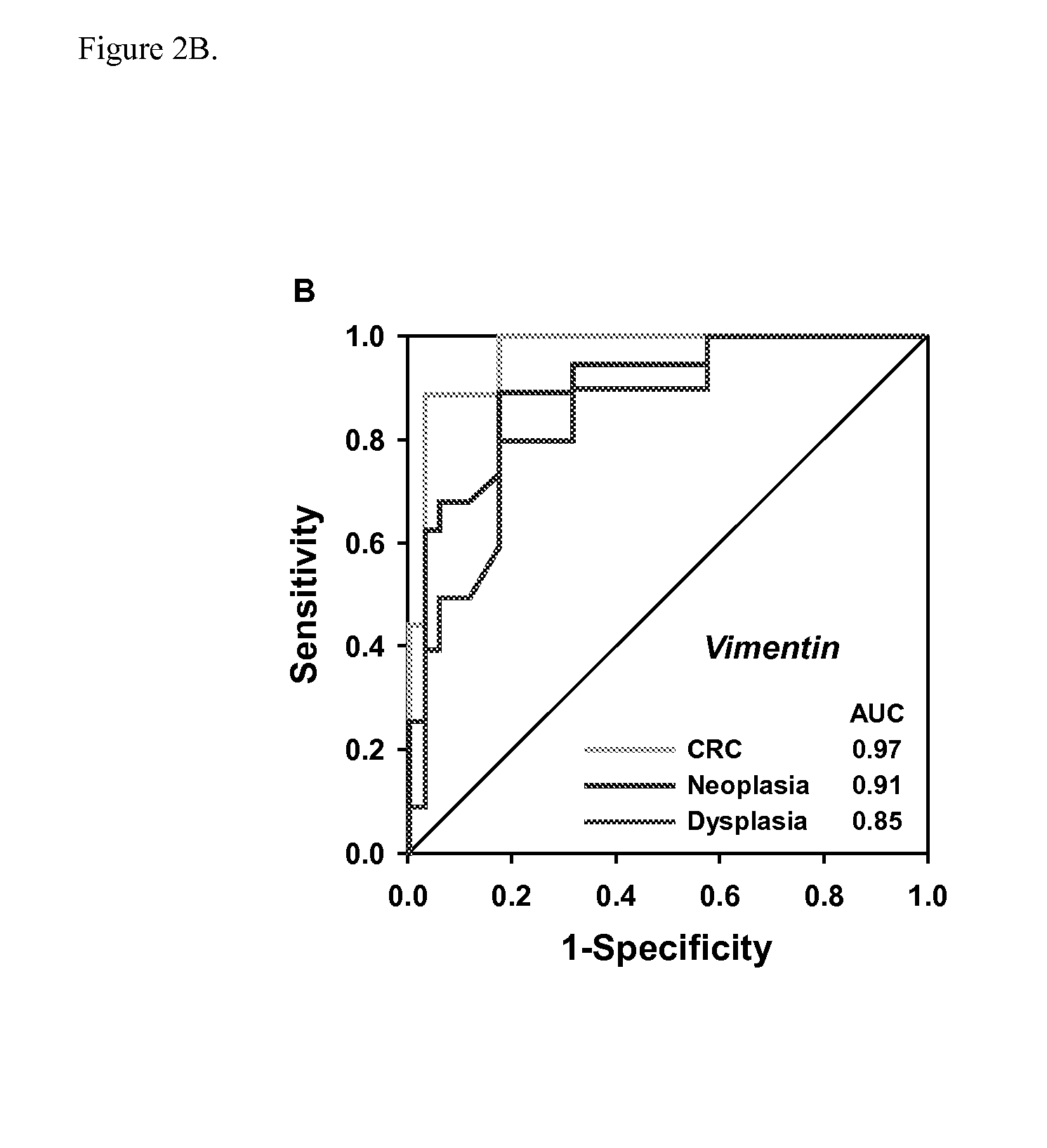
The present invention provides methods and materials related to the detection of colorectal neoplasm-specific markers (e.g., markers associated with colorectal cancer, markers associated with adenoma) in or associated with a subject's stool sample. In particular, the present invention provides methods and materials for identifying mammals (e.g., humans) having a colorectal neoplasm by detecting the presence and level of indicators of colorectal neoplasia such as, for example, long DNA (e.g., quantified by Alu PCR) and the presence and level of tumor-associated gene alterations (e.g., mutations in KRAS, APC, melanoma antigen gene, p53, BRAF, BAT26, PIK3CA) or epigenetic alterations (e.g., DNA methylation) (e.g., CpG methylation) (e.g., CpG methylation in coding or regulatory regions of bmp-3, bmp-4, SFRP2, vimentin, septin9, ALX4, EYA4, TFPI2, NDRG4, FOXE1) in DNA from a stool sample obtained from the mammal.
Owner:MAYO FOUND FOR MEDICAL EDUCATION & RES
Regulation of epigenetic control of gene expression
Methods are provided for the identification of compounds that selectively modulate epigenetic changes in gene expression. Compounds, compositions, kits or assays devices, and methods are provided for modulating the expression, endogenous levels or the function of small non-coding RNAs cognate to or transcribed by heterochromatic regions subject to epigenetic regulation (i.e., promoters, enhancers, centromeres, telomeres, origins of DNA replication, imprinted loci, or loci marked by dosage-compensation), and for modulating the formation or function of heterochromatin in cells, tissues or animals.
Owner:IONIS PHARMA INC
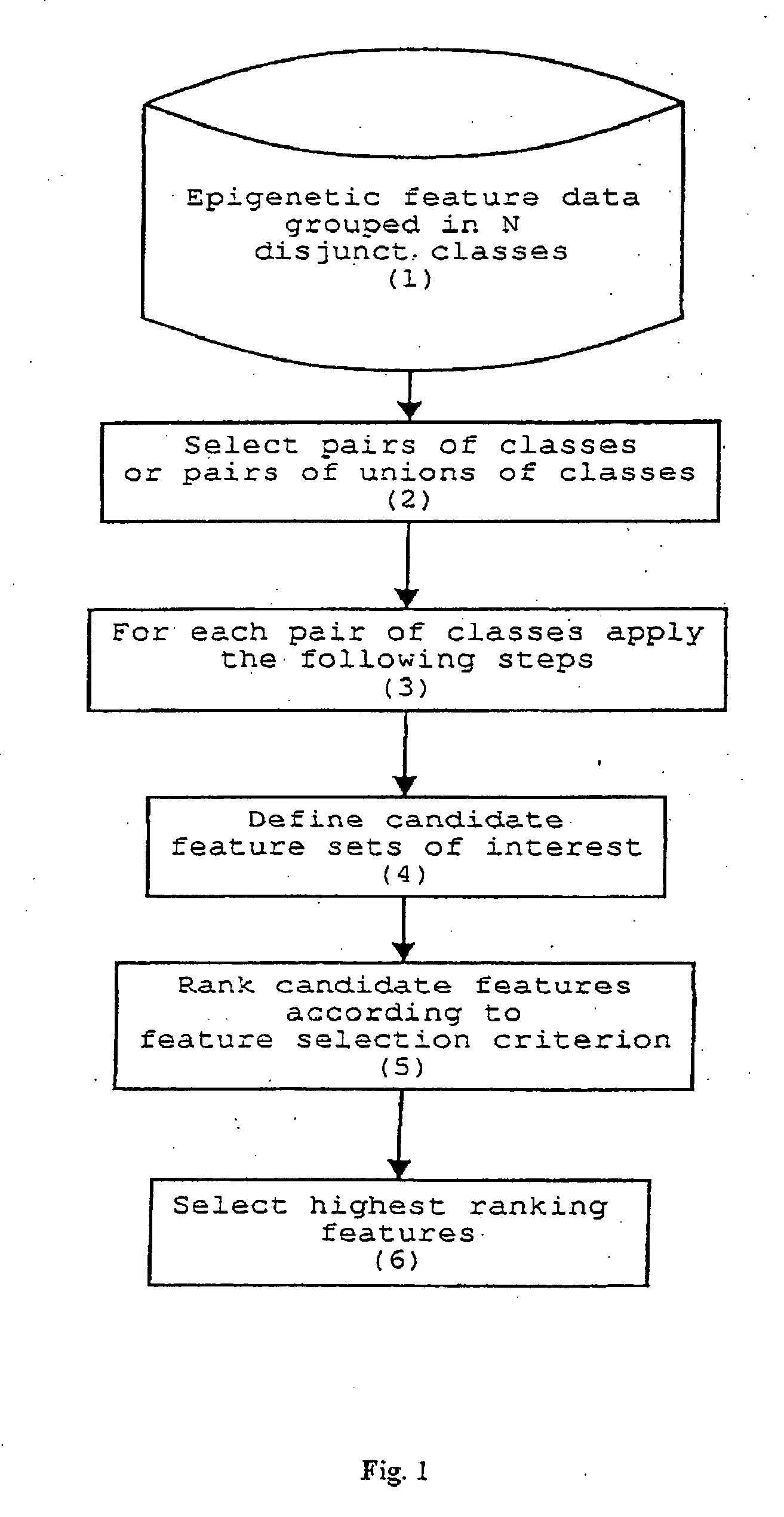
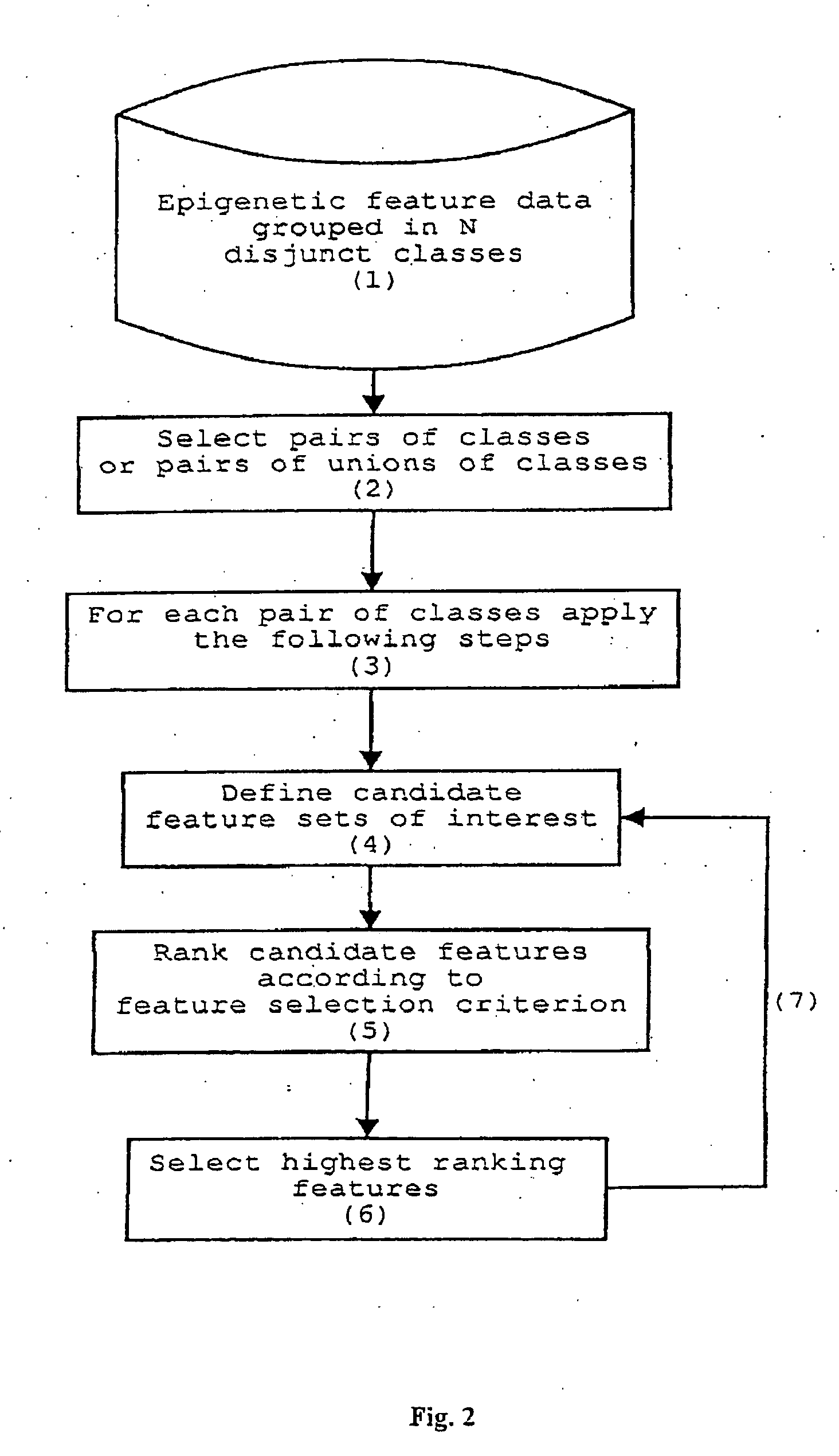
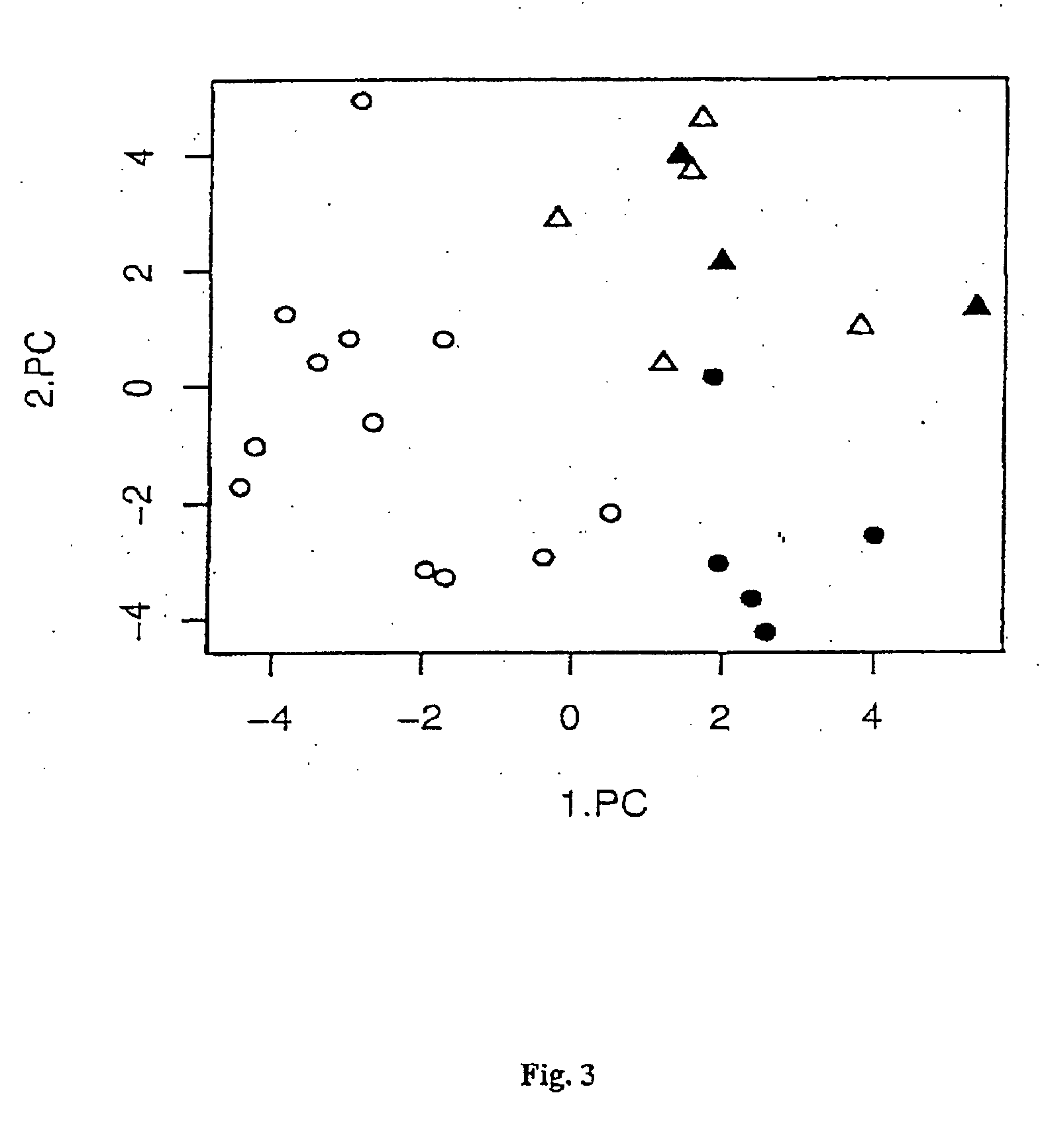
Method for epigenetic feature selection
InactiveUS20040102905A1Improve performanceMicrobiological testing/measurementBiological testingData setEpigenetic Profile
A method for selecting epigenetic features includes receiving an epigenetic feature data set for a plurality of epigenetic features of interest. The epigenetic feature data set is grouped in disjunct classes of interest. Epigenetic features of interest and / or combinations of epigenetic features of interest are selected that are relevant for epigenetically-based prediction based on corresponding epigenetic feature data. A new set of epigenetic features of interest is defined based on the relevant epigenetic features of interest and / or combinations of epigenetic features of interest.
Owner:EPIGENOMICS AG
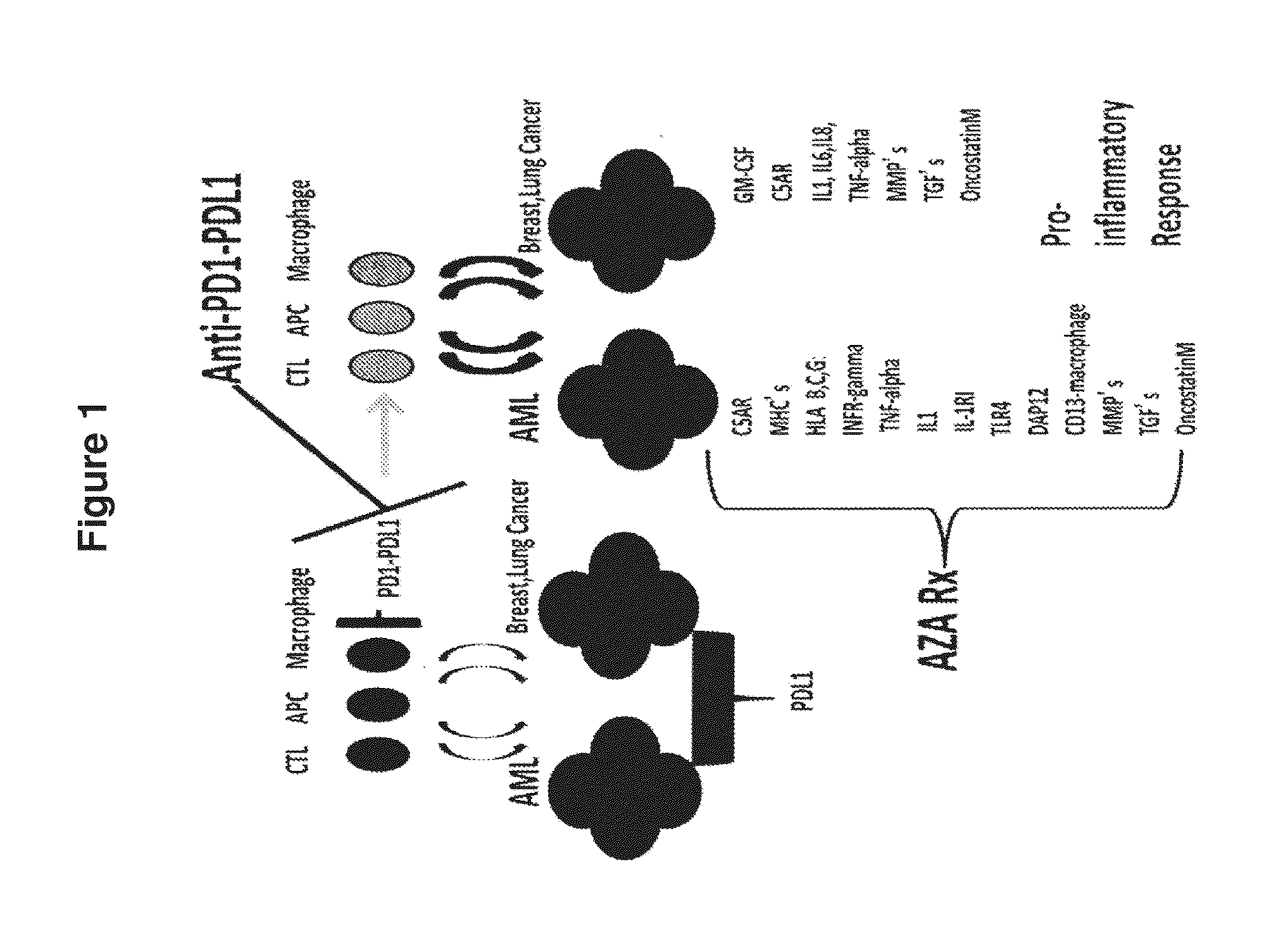
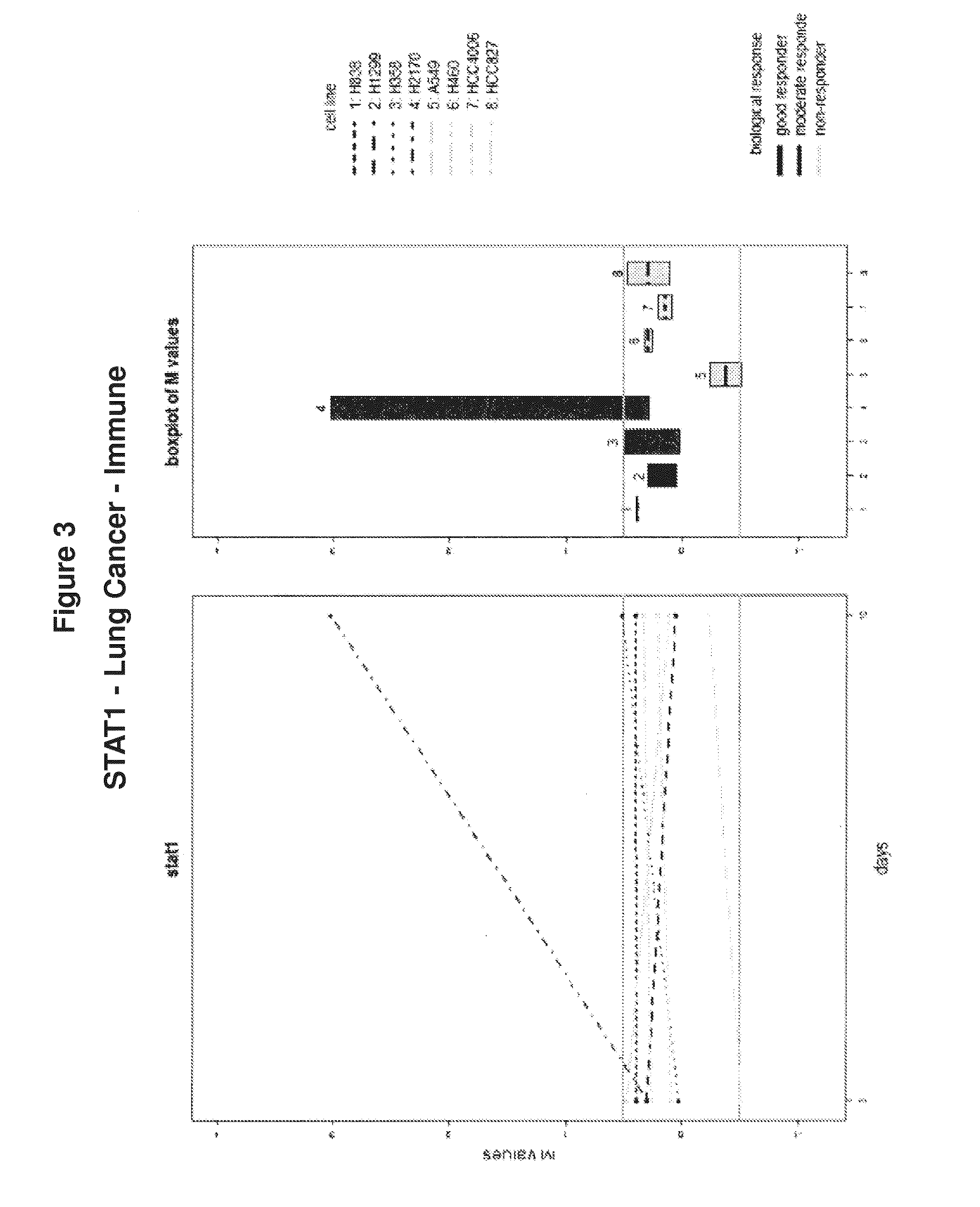
Cancer therapy via a combination of epigenetic modulation and immune modulation
ActiveUS20160193239A1Improve immunityAnti-neoplastic effect of the immune modulating agent in the subject is enhancedOrganic active ingredientsMicrobiological testing/measurementAntiendomysial antibodiesImmune modulator
Cancer therapies that combine epigenetic modulating agent(s) with immune modulating agent(s), which were remarkably identified to provide an improved treatment regimen over single agent therapy, are disclosed. In particular embodiments, the invention provides for improved treatment of NSCLC in patients via administration of exemplary immune modulating agents anti-PD-1 antibody or anti-PD-L1 antibody, which were observed to show enhanced activity in combination with the exemplary epigenetic modulating agent 5-deoxyazacytidine. Further, expression markers of responsive neoplastic cells are also disclosed.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Diagnosis of diseases associated with dna transcription
The present invention relates to the chemically modified genomic sequences of genes associated with DNA transcription to oligonucleotides and / or PNA-oligomers for detecting the cytosine methylation state of genes associated with DNA transcription which are directed against the sequence as well as to a method for ascertaining genetic and / or epigenetic parameters of genes associated with DNA transcription.
Owner:EPIGENOMICS AG
Method and nucleic acids for the analysis of astrocytomas
InactiveUS20040115630A1Preferred pairing propertyEasy to detectBioreactor/fermenter combinationsBiological substance pretreatmentsCytosineOligomer
Owner:EPIGENOMICS AG
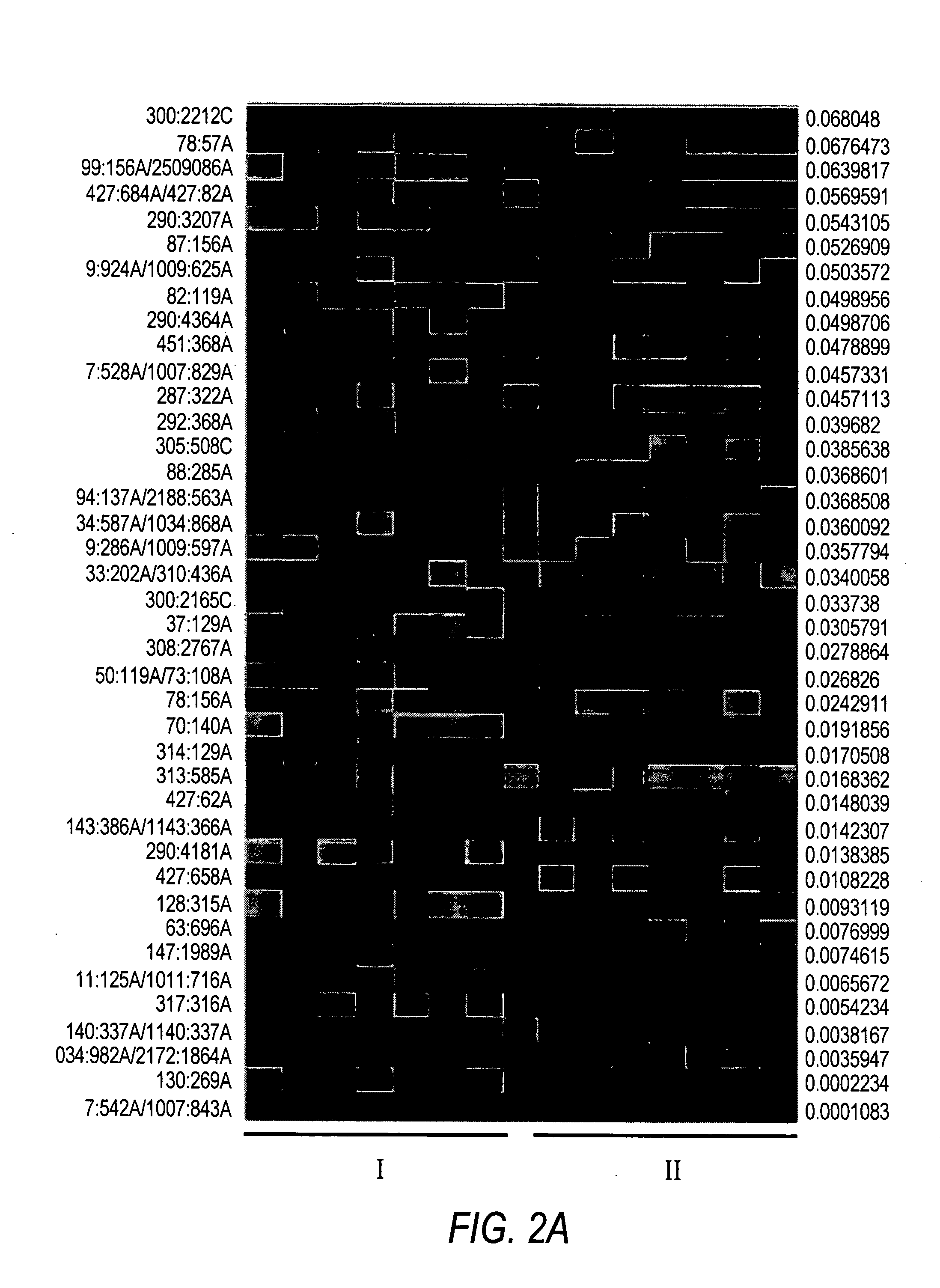
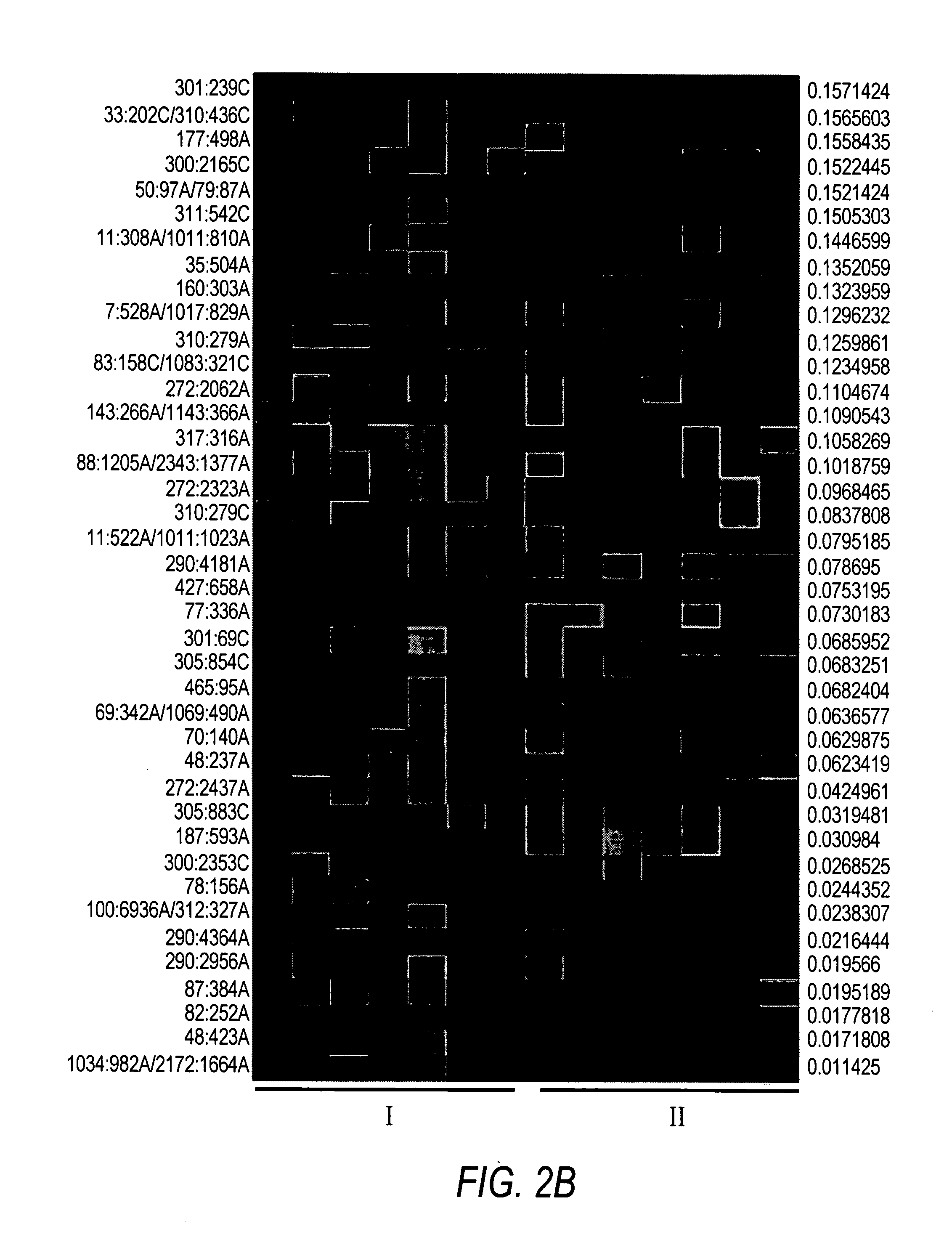
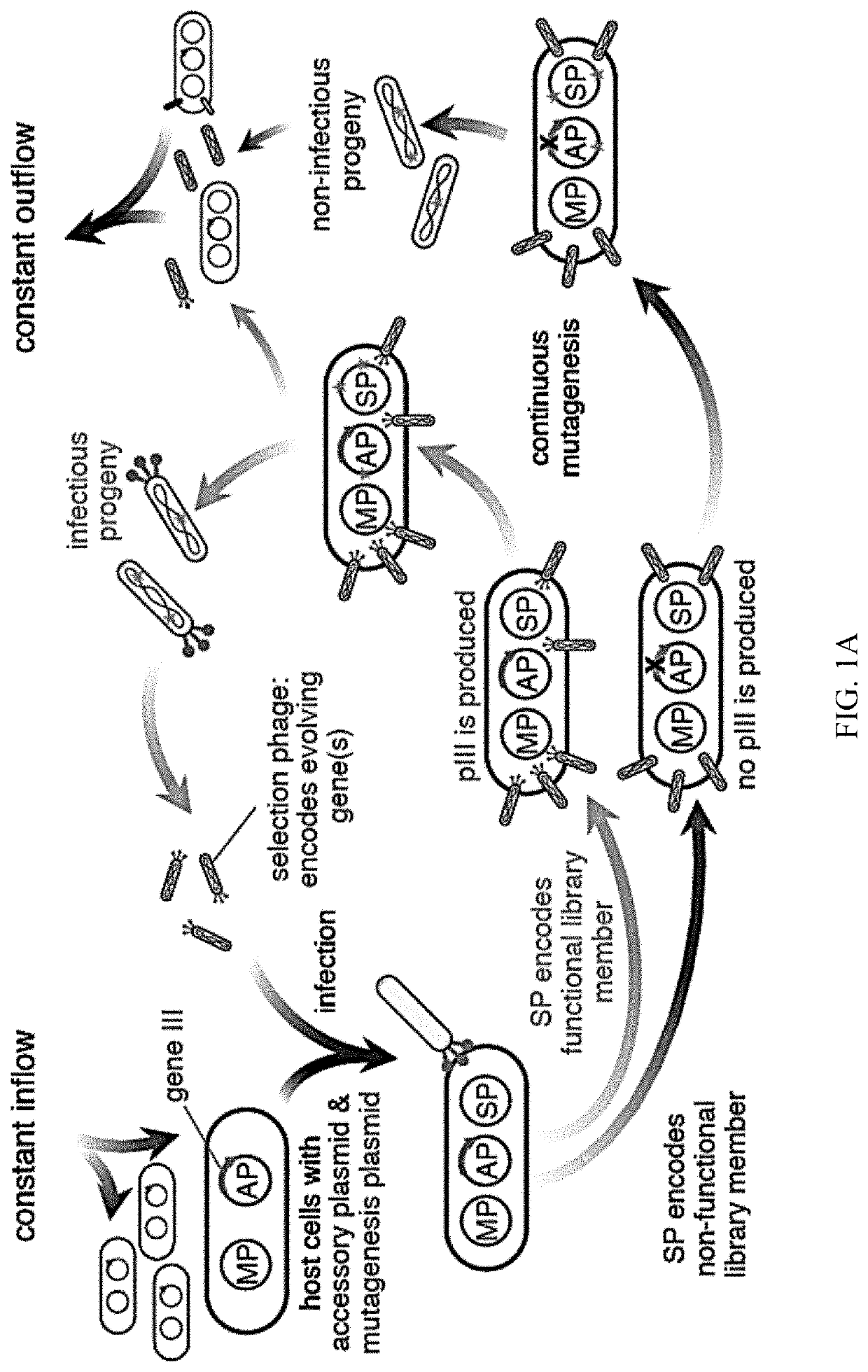
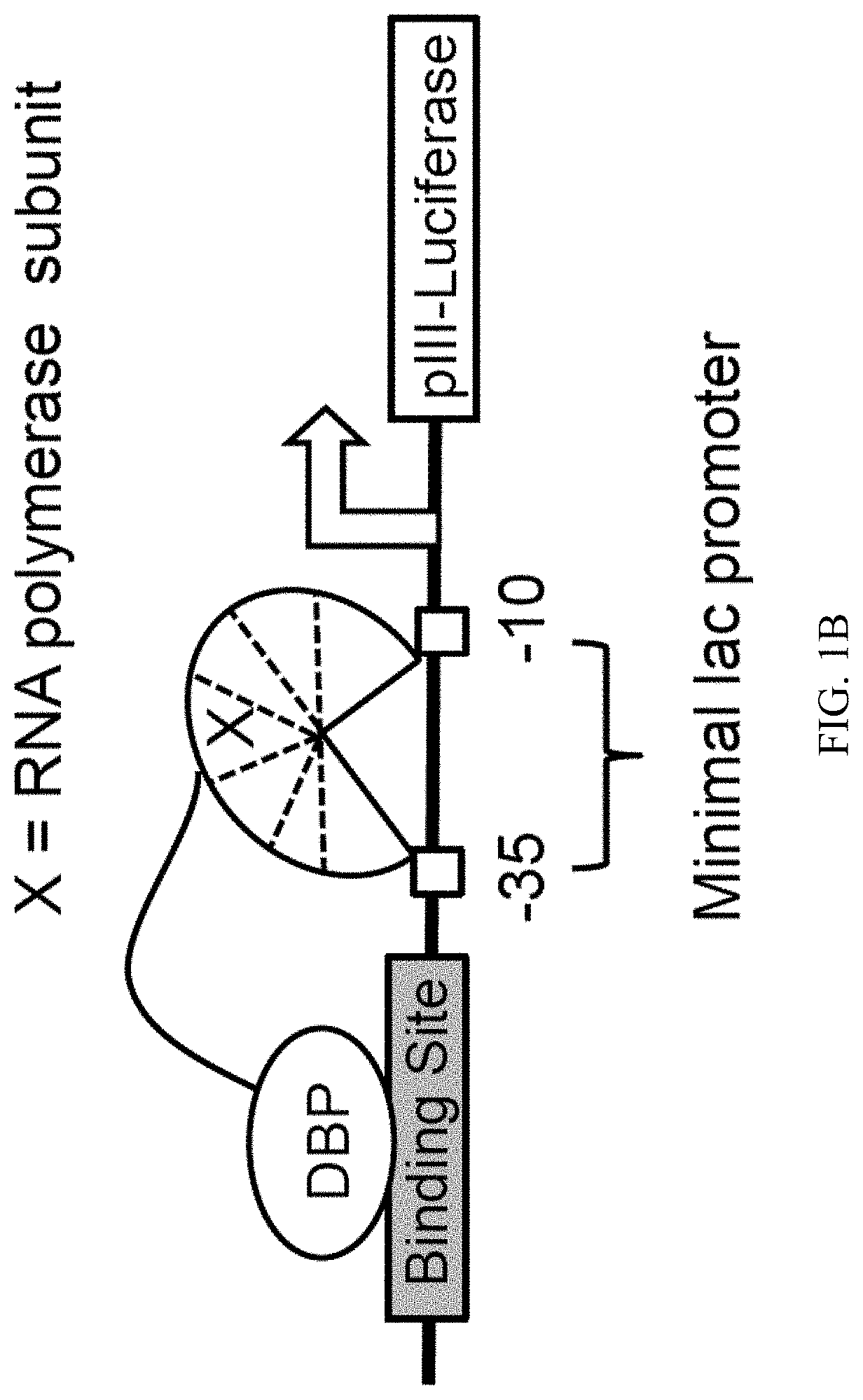
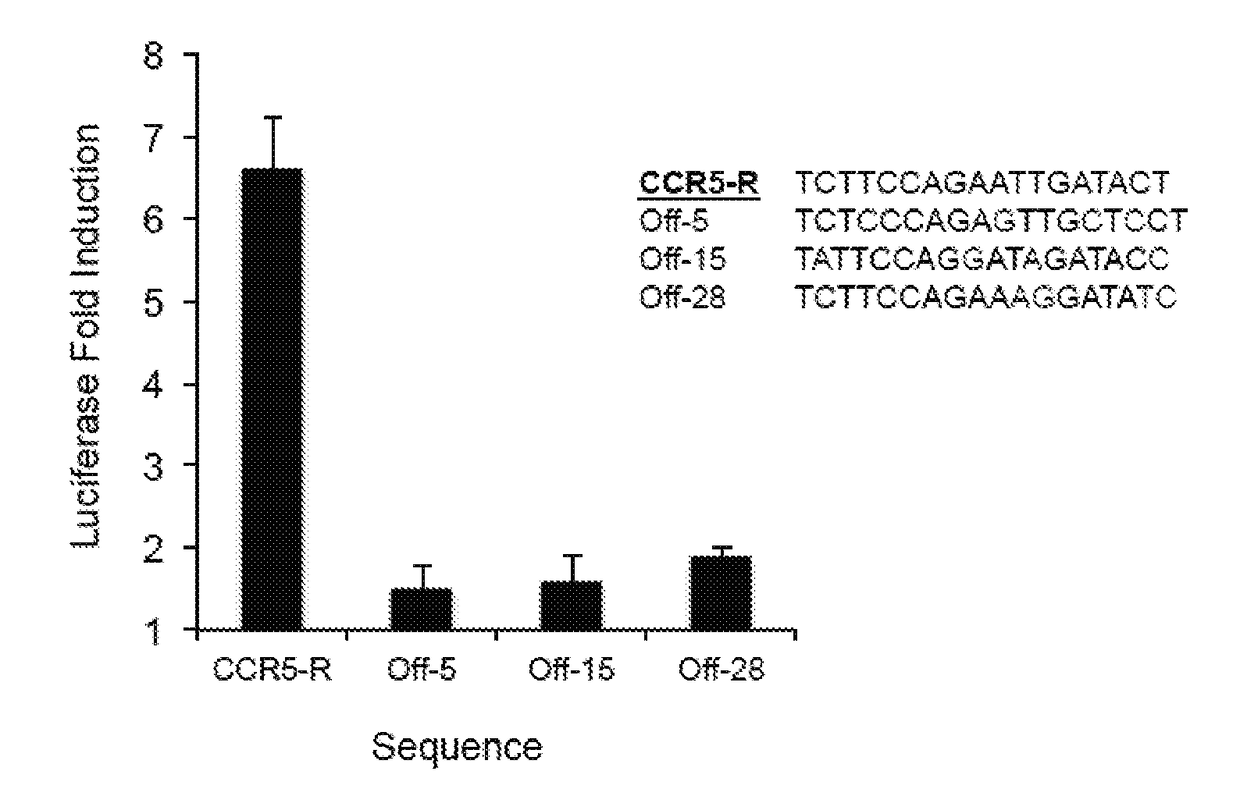
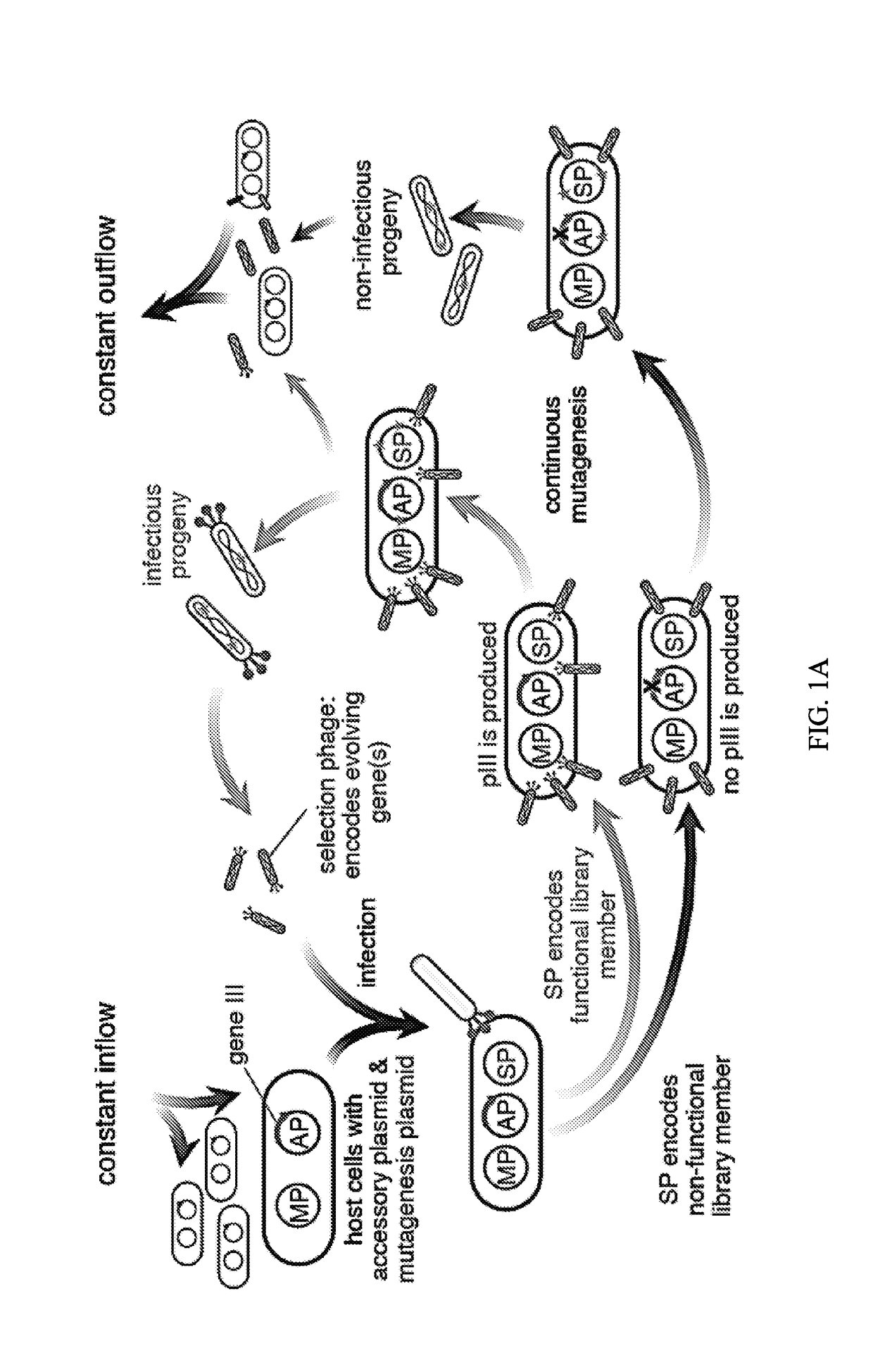
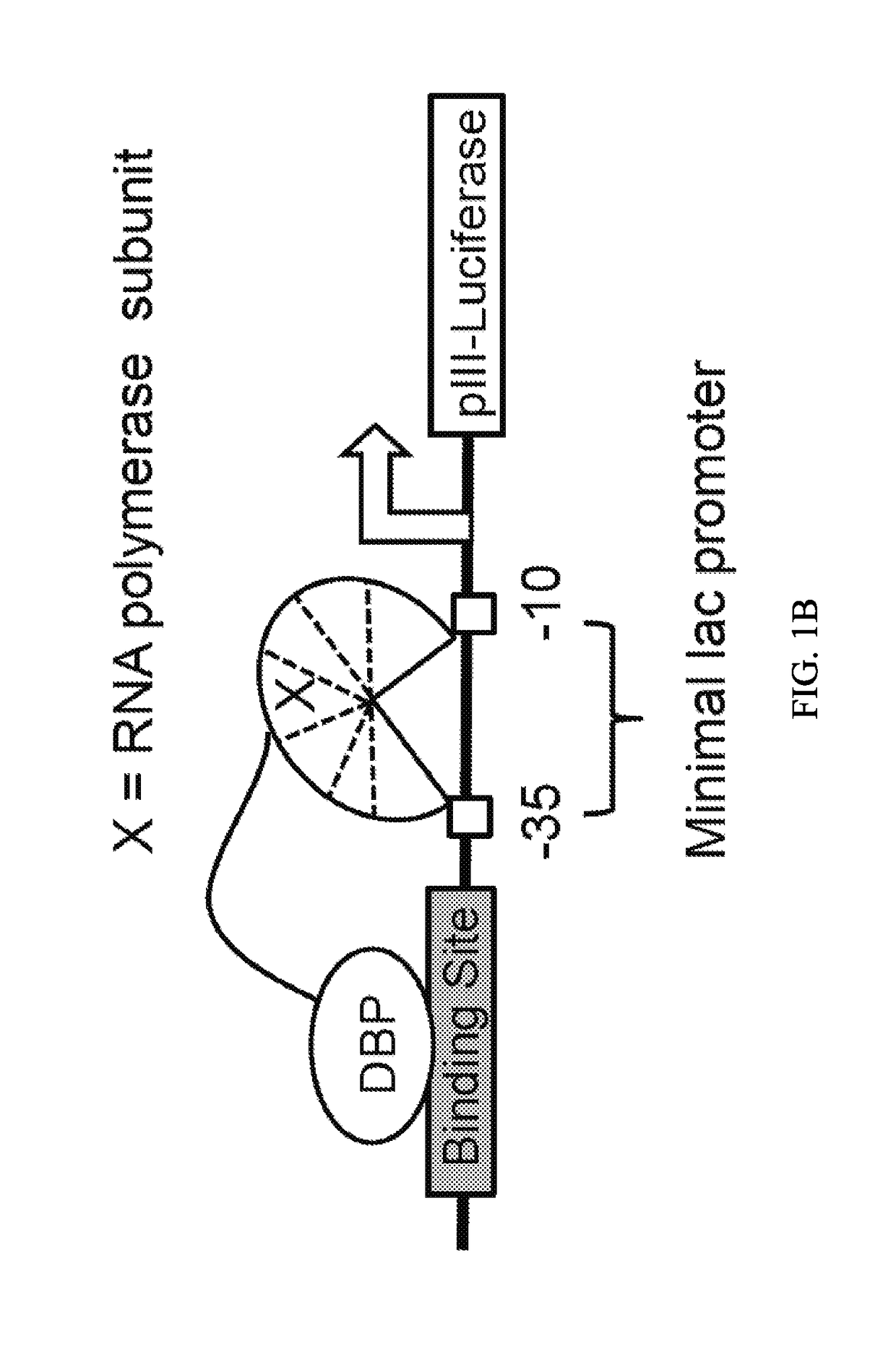
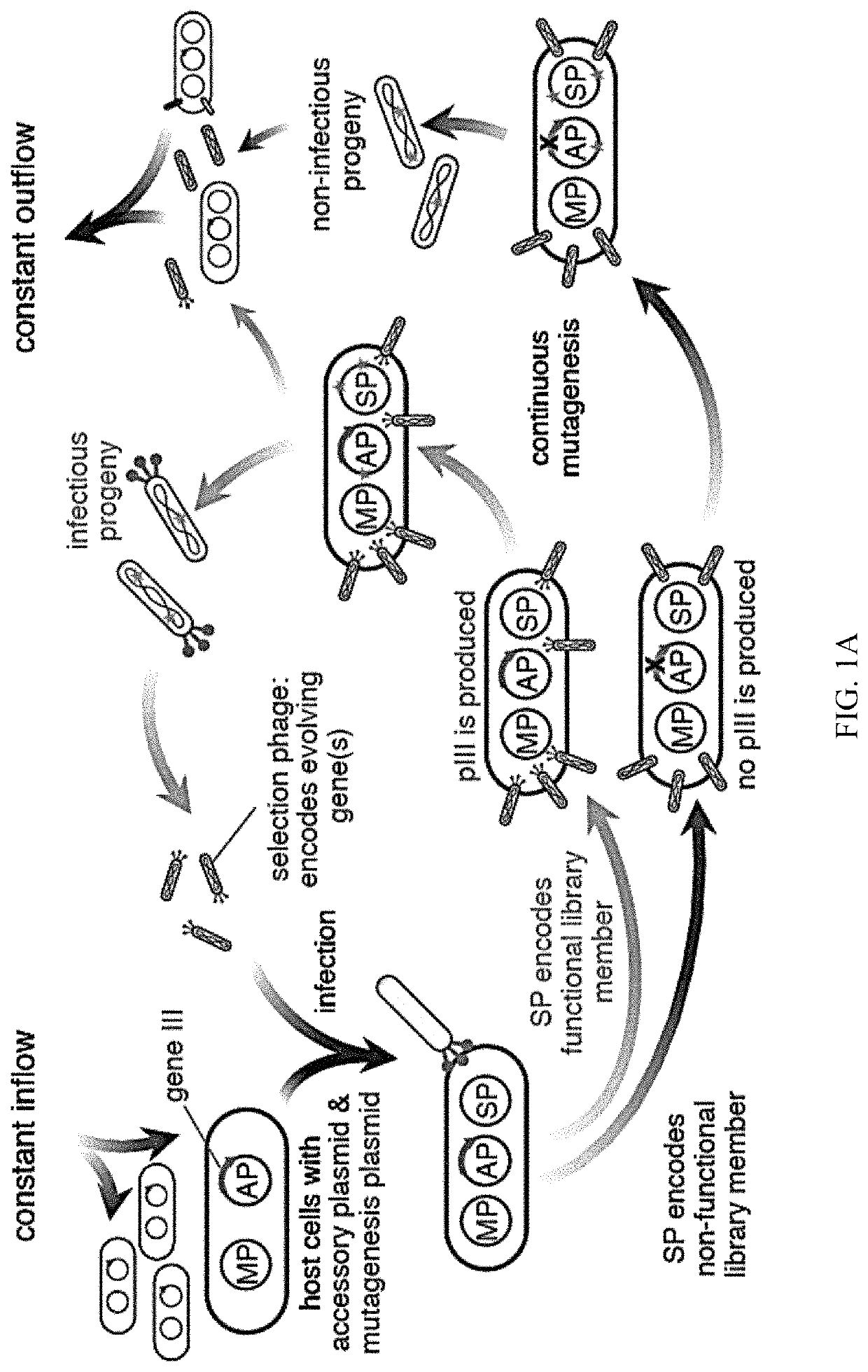
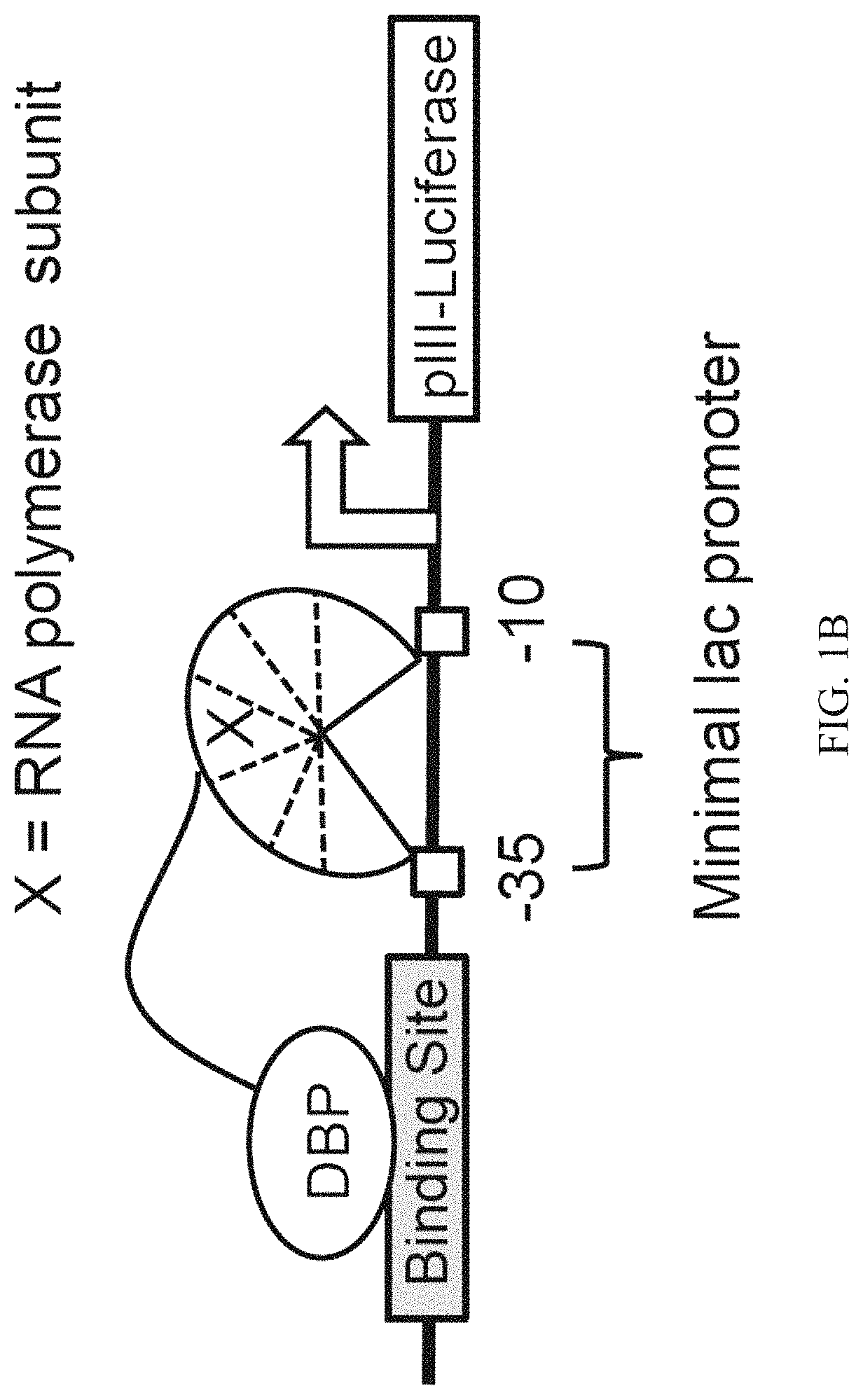
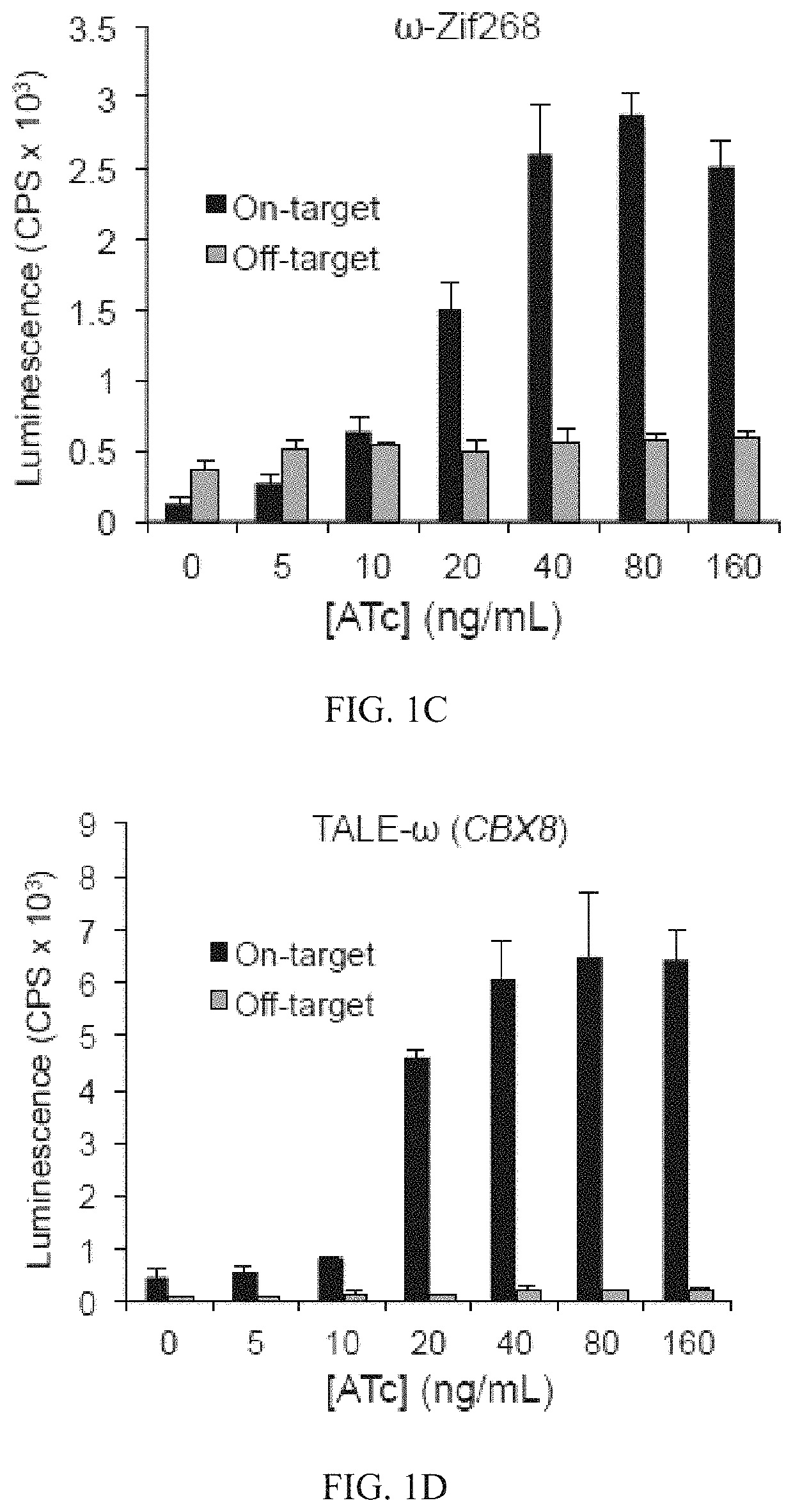
Evolution of talens
ActiveUS20200277587A1Strong specificityHydrolasesGenetic therapy composition manufactureNucleotideEpigenetic Profile
Engineered transcriptional activator-like effectors (TALEs) are versatile tools for genome manipulation with applications in research and clinical contexts. One current drawback of TALEs is that the 5′ nucleotide of the target is specific for thymine (T). TALE domains with alternative 5′ nucleotide specificities could expand the scope of DNA target sequences that can be bound by TALEs. Another drawback of TALEs is their tendency to bind and cleave off-target sequence, which hampers their clinical application and renders applications requiring high-fidelity binding unfeasible. This disclosure provides methods and strategies for the continuous evolution of proteins comprising DNA-binding domains, e.g., TALE domains. In some aspects, this disclosure provides methods and strategies for evolving such proteins under positive selection for a desired DNA-binding activity and / or under negative selection against one or more undesired (e.g., off-target) DNA-binding activities. Some aspects of this disclosure provide engineered TALE domains and TALEs comprising such engineered domains, e.g., TALE nucleases (TALENs), TALE transcriptional activators, TALE transcriptional repressors, and TALE epigenetic modification enzymes, with altered 5′ nucleotide specificities of target sequences. Engineered TALEs that target ATM with greater specificity are also provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Methods of detecting colorectal cancer
InactiveUS20120164238A1Prevents denaturation and degradationBiocideInorganic active ingredientsColorectal cancerOncology
A method of detecting a predisposition to, or the incidence of colorectal cancer in a faecal sample comprises, in a first step (a), detecting the presence of blood in the faecal sample, wherein detection of the presence of blood is indicative of a predisposition to, or the incidence of colorectal cancer. The method additionally comprises, in second step (b), detecting an epi-genetic modification in the DNA contained within the faecal sample, wherein detection of the epigenetic modification is indicative of a predisposition to, or the incidence of colorectal cancer. Based upon a positive result obtained in either (a) or (b) or in both (a) and (b) a predisposition to, or the incidence of colorectal cancer is detected. Related methods and kits involve detecting an epigenetic modification in a number of specific genes.
Owner:EXACT SCI CORP
Modifiable attribute identification
ActiveUS8788283B2Level of resolutionHigh combinationPhysical therapies and activitiesDigital data processing detailsLongevityData science
Owner:23ANDME
Evolution of talens
ActiveUS20180237758A1Strong specificityHydrolasesGenetic therapy composition manufactureDNA-binding domainOff targets
Engineered transcriptional activator-like effectors (TALEs) are versatile tools for genome manipulation with applications in research and clinical contexts. One current drawback of TALEs is that the 5′ nucleotide of the target is specific for thymine (T). TALE domains with alternative 5′ nucleotide specificities could expand the scope of DNA target sequences that can be bound by TALEs. This disclosure provides methods and strategies for the continuous evolution of proteins comprising DNA-binding domains, e.g., TALE domains. In some aspects, this disclosure provides methods and strategies for evolving such proteins under positive selection for a desired DNA-binding activity and / or under negative selection against one or more undesired (e.g., off-target) DNA-binding activities. Some aspects of this disclosure provide engineered TALE domains and TALEs comprising such engineered domains, e.g., TALE nucleases (TALENs), TALE transcriptional activators, TALE transcriptional repressors, and TALE epigenetic modification enzymes, with altered 5′ nucleotide specificities of target sequences. Engineered TALEs that target ATM with greater specificity are also provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
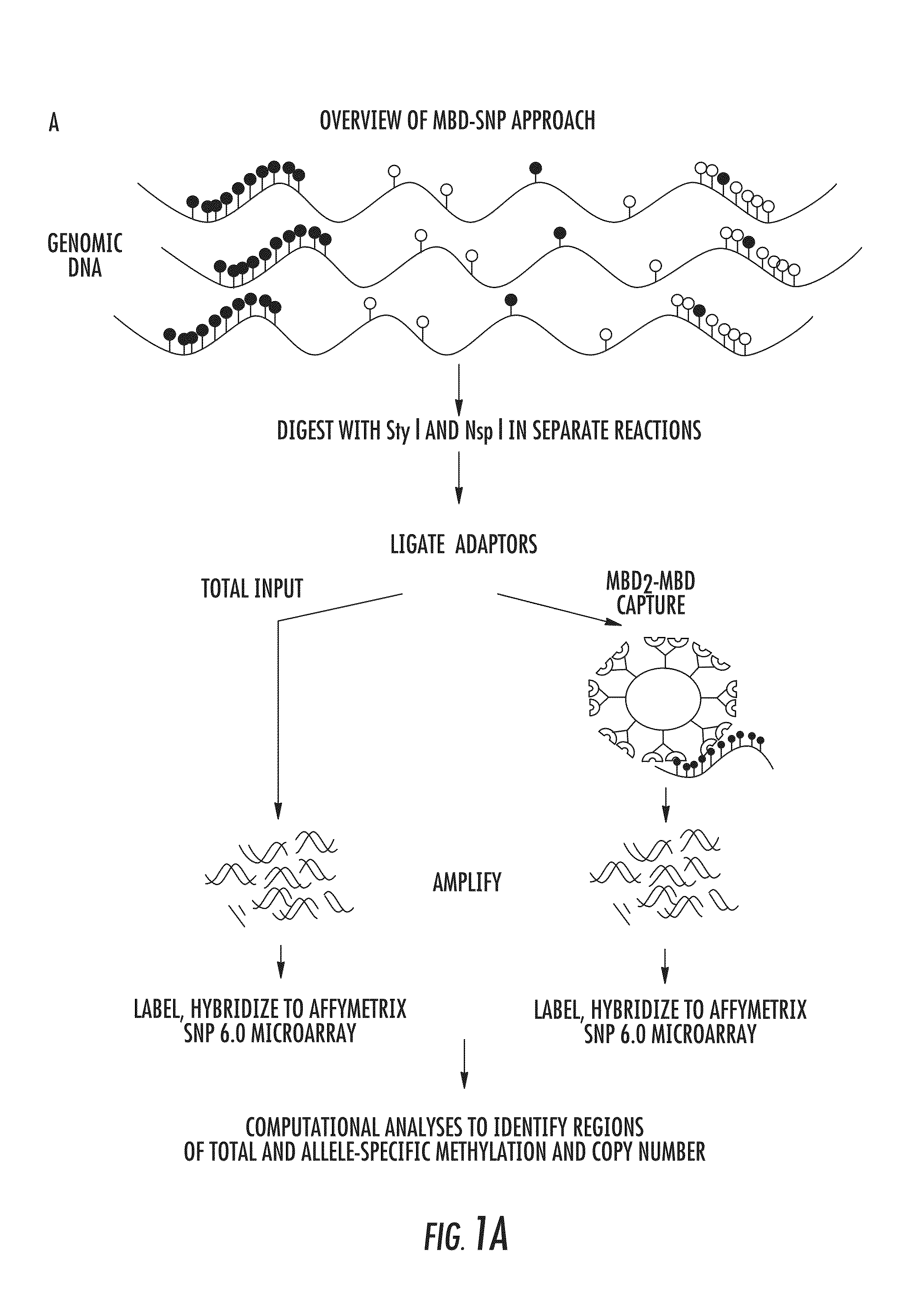
DNA methylation markers for metastatic prostate cancer
InactiveUS20140274767A1Rapid and cost-effectiveMicrobiological testing/measurementLibrary screeningDNA methylationGenome scale
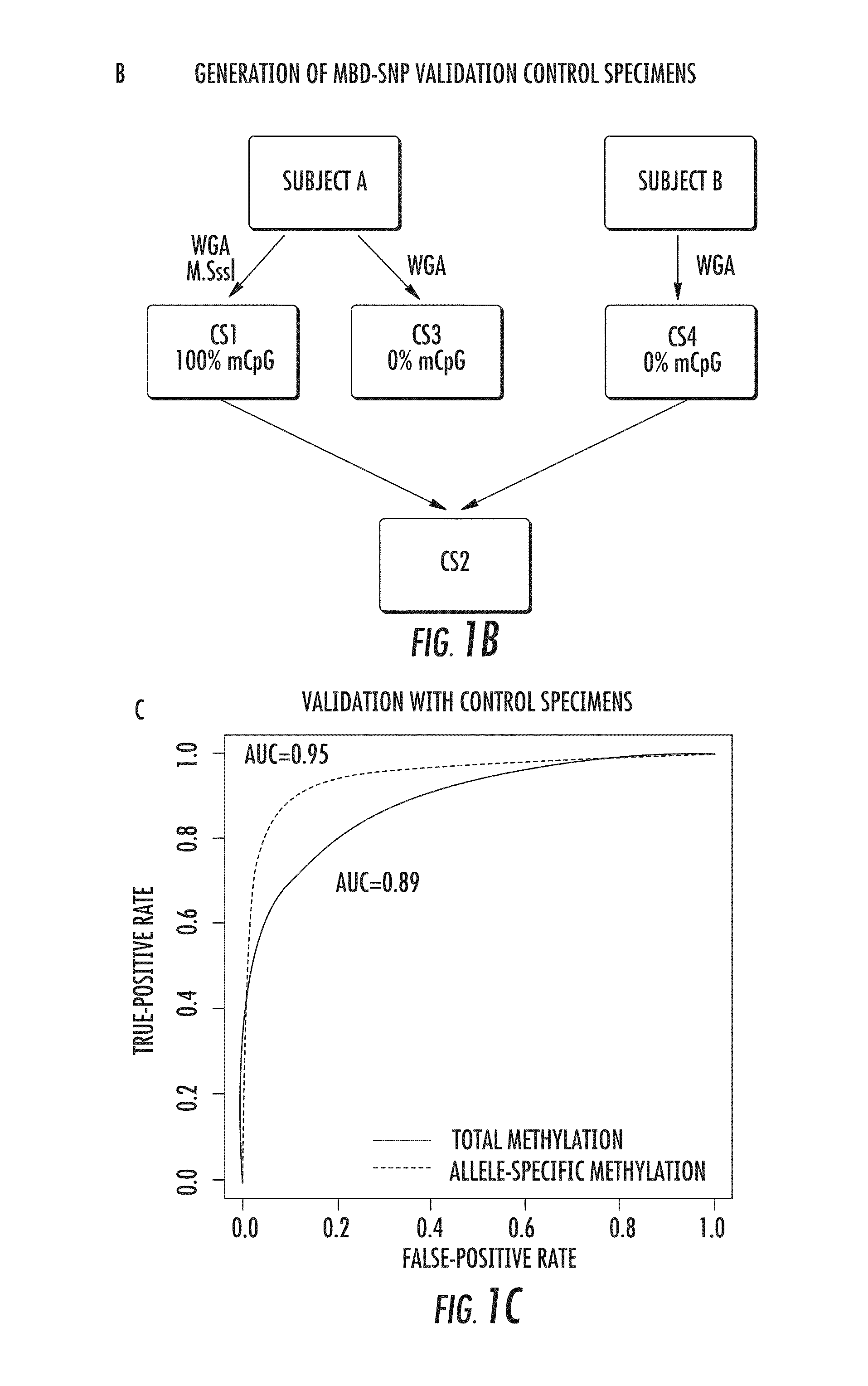
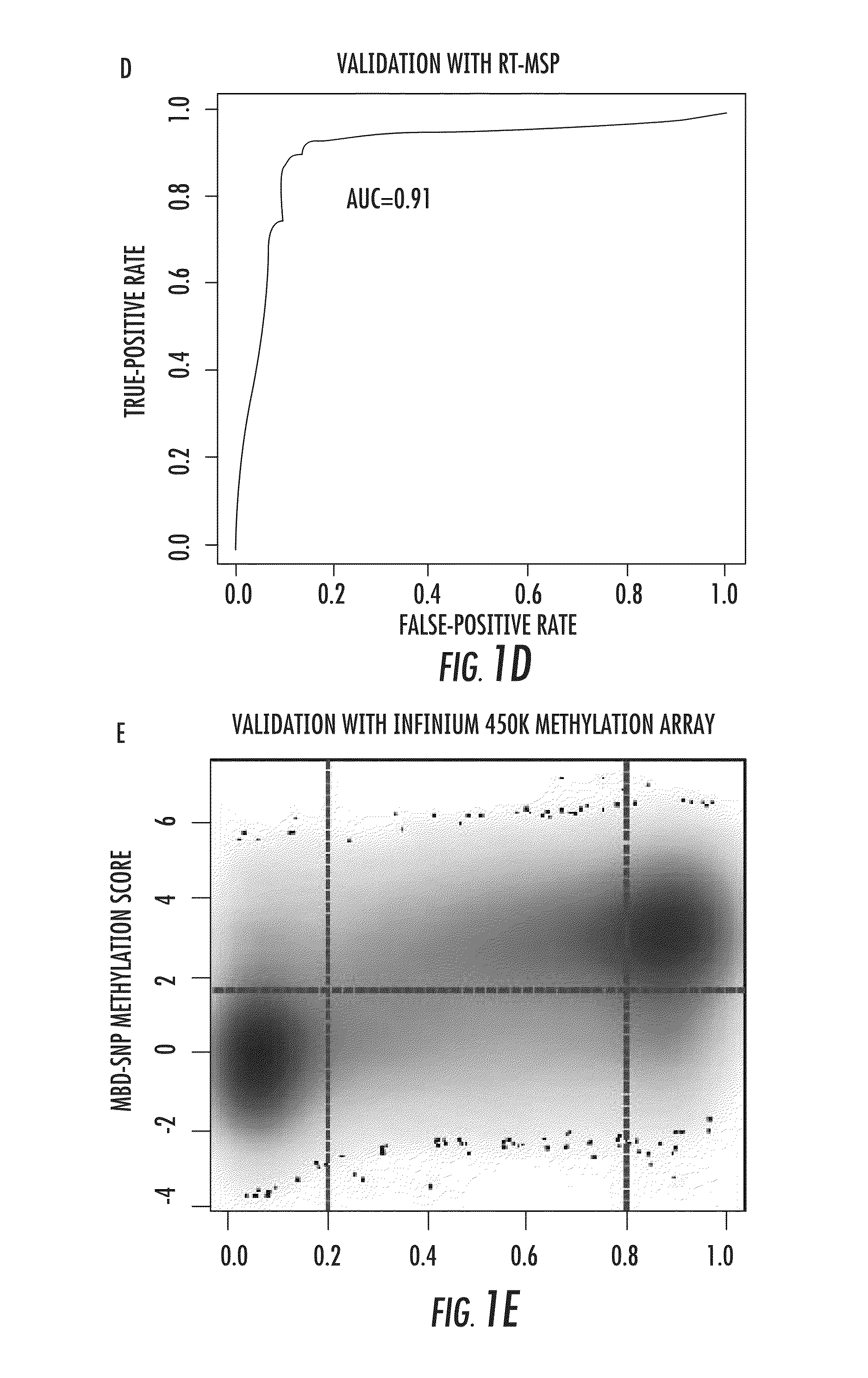
The present invention relates to the field of cancer. More specifically, the present invention provides methods and compositions useful for assessing prostate cancer. In a specific embodiment, present inventors have developed and applied a new technology and associated computation methods enabling simultaneous genome-scale analysis of genetic (copy number) and epigenetic (total methylation (TM) and allele-specific methylation (ASM) alternation, This method, called MBD-SNP, features affinity enrichment or methylated genomic DNA fragments using a methyl-binding domain polypeptide.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Systems, Kits, and Tests for Detecting Colorectal Cancer
InactiveUS20160194723A1Prevents denaturation and degradationMicrobiological testing/measurementDisease diagnosisEpigenetic ProfileOncology
A method of detecting a predisposition to, or the incidence of, colorectal cancer in a fecal sample comprises, in a first step (a), detecting the presence of blood in the fecal sample, wherein detection of the presence of blood is indicative of a predisposition to, or the incidence of, colorectal cancer. The method additionally comprises, in second step (b), detecting an epigenetic modification in the DNA contained within the fecal sample, wherein detection of the epigenetic modification is indicative of a predisposition to, or the incidence of, colorectal cancer. Based upon a positive result obtained in either (a) or (b) or in both (a) and (b) a predisposition to, or the incidence of, colorectal cancer is detected. Related methods and kits involve detecting an epigenetic modification in a number of specific genes.
Owner:EXACT SCI CORP
Administration Of DNA Methylation Inhibitors For Treating Epigenetic Diseases
Methods are provided for treating patients with epigenetic diseases, especially those associated with aberrant DNA methylation such as hematological disorders and cancer. By administering a DNA methylation inhibitor to the patients following unique dosing regimens, the disease can be efficaciously treated with reduced toxic side effects.
Owner:SUPERGEN
Methods and materials for noninvasive detection of colorectal neoplasia associated with inflammatory bowel disease
InactiveUS20130244235A1Improve discriminationSensitive highMicrobiological testing/measurementDNA methylationMammal
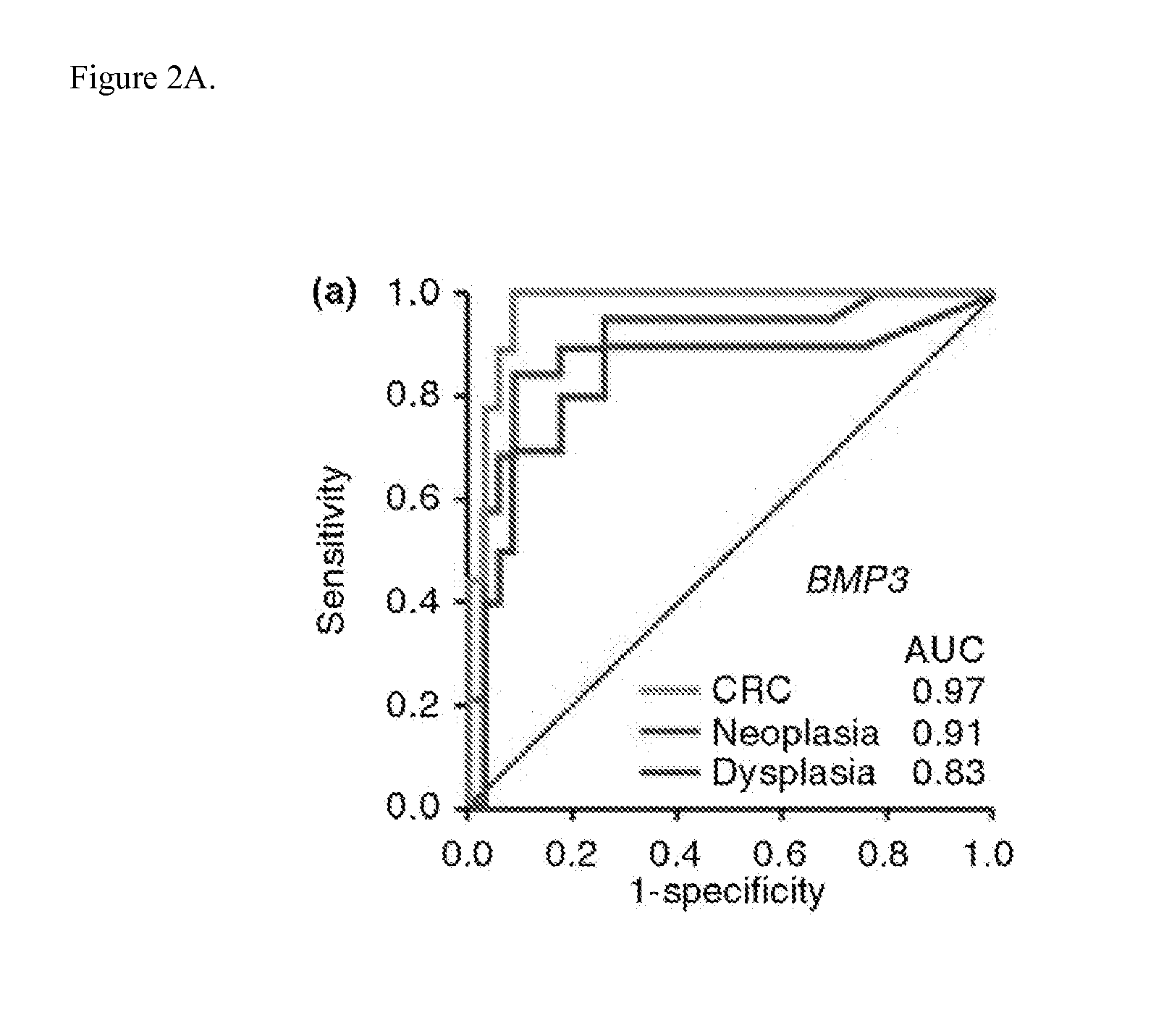
The present invention provides methods and materials related to the detection of colorectal neoplasia (CRN) associated with inflammatory bowel disease (IBD). The present invention provides markers specific for colorectal neoplasia associated with inflammatory bowel disease in or associated with a subject's stool sample. In particular, the present invention provides methods and materials for identifying mammals (e.g., humans) having colorectal neoplasia associated with inflammatory bowel disease by detecting the presence and level of indicators of colorectal neoplasia such as, for example, epigenetic alterations (e.g., DNA methylation) (e.g., CpG methylation) (e.g., CpG methylation in coding or regulatory regions of BMP3, NDRG4, vimentin, EYA4) in DNA from a stool sample obtained from the mammal.
Owner:MAYO FOUND FOR MEDICAL EDUCATION & RES
Evolution of TALENs
ActiveUS10612011B2Strong specificityHydrolasesGenetic therapy composition manufactureNucleotideEpigenetic Profile
Engineered transcriptional activator-like effectors (TALEs) are versatile tools for genome manipulation with applications in research and clinical contexts. One current drawback of TALEs is that the 5′ nucleotide of the target is specific for thymine (T). TALE domains with alternative 5′ nucleotide specificities could expand the scope of DNA target sequences that can be bound by TALEs. This disclosure provides methods and strategies for the continuous evolution of proteins comprising DNA-binding domains, e.g., TALE domains. In some aspects, this disclosure provides methods and strategies for evolving such proteins under positive selection for a desired DNA-binding activity and / or under negative selection against one or more undesired (e.g., off-target) DNA-binding activities. Some aspects of this disclosure provide engineered TALE domains and TALEs comprising such engineered domains, e.g., TALE nucleases (TALENs), TALE transcriptional activators, TALE transcriptional repressors, and TALE epigenetic modification enzymes, with altered 5′ nucleotide specificities of target sequences. Engineered TALEs that target ATM with greater specificity are also provided.
Owner:PRESIDENT & FELLOWS OF HARVARD COLLEGE
Diagnosis of diseases associated with angiogenesis
The present invention concerns the chemically modified genomic sequences of genes associated with angiogenesis, oligonucleotides and / or PNA oligomers directed against the sequence for the detection of the cytosine methylation state of genes associated with angiogenesis as well as a method for determining genetic and / or epigenetic parameters of genes associated with angiogenesis.
Owner:EPIGENOMICS AG
Method and nucleic acids for the differentiation of prostate tumors
InactiveUS20030113750A1Preferred pairing propertyEasy to detectBioreactor/fermenter combinationsOrganic active ingredientsCytosineGenomic DNA
The present invention relates to chemically modified genomic sequences, to oligonucleotides and / or PNA-oligomers for detecting the cytosine methylation state of genomic DNA, as well as to a method for ascertaining genetic and / or epigenetic parameters of genes for use in characterisation, classification, differentiation, diagnosis and therapy of prostate lesions.
Owner:EPIGENOMICS AG
Methods of diagnosing cancer using epigenetic biomarkers
InactiveUS20140213475A1Easy to quantifyEasy to distinguishMicrobiological testing/measurementLibrary screeningCancer cellEpigenetic Profile
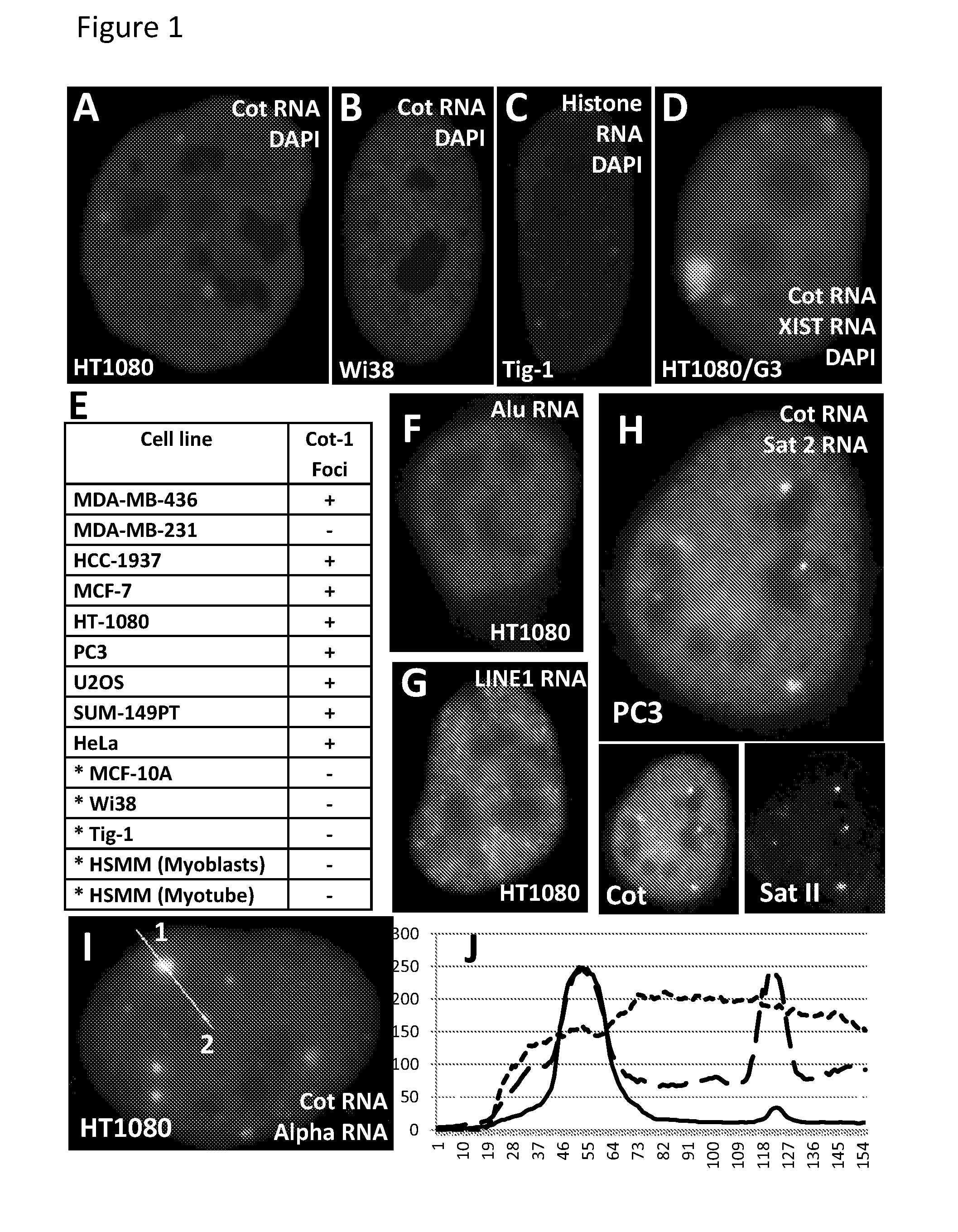
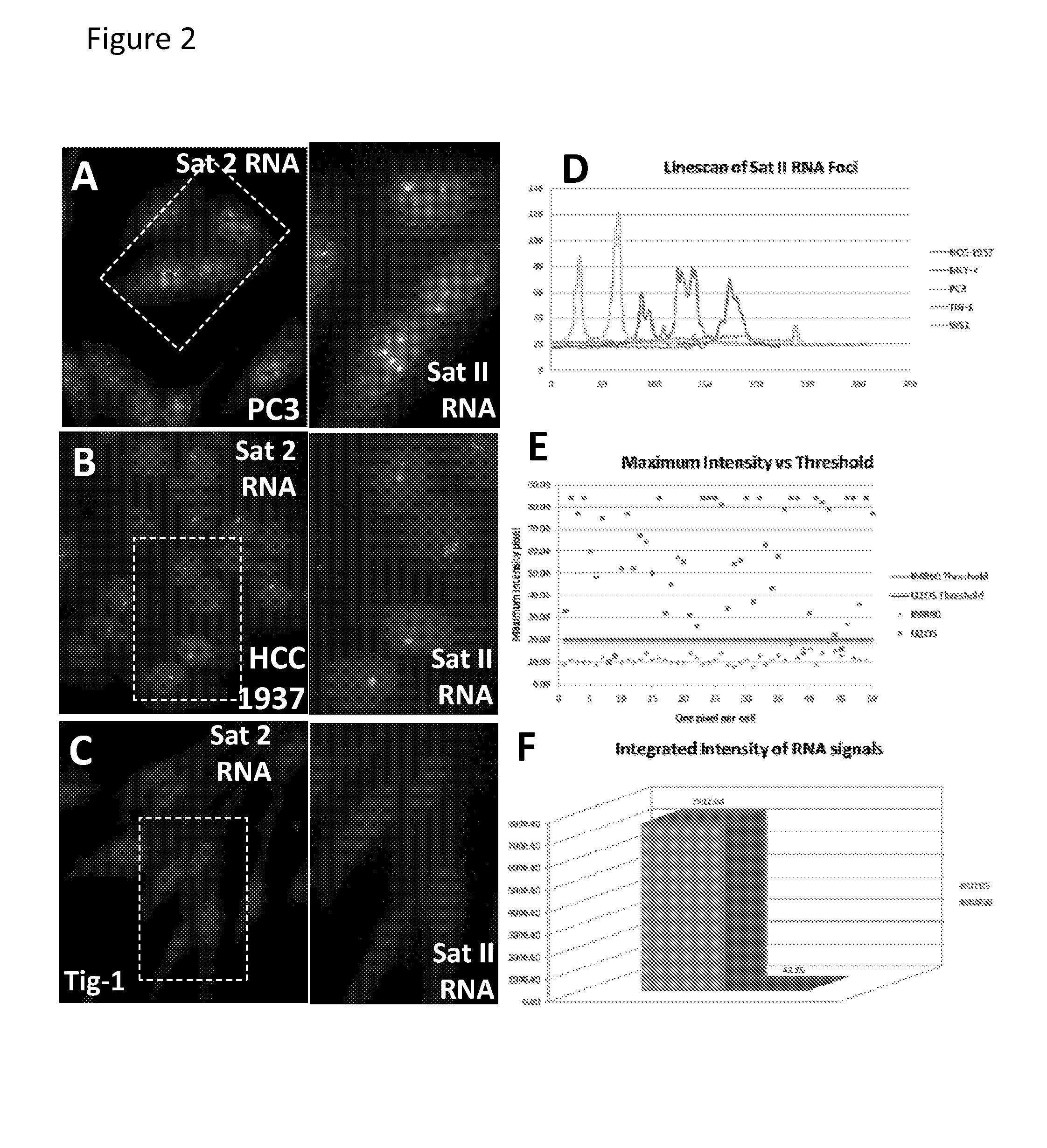
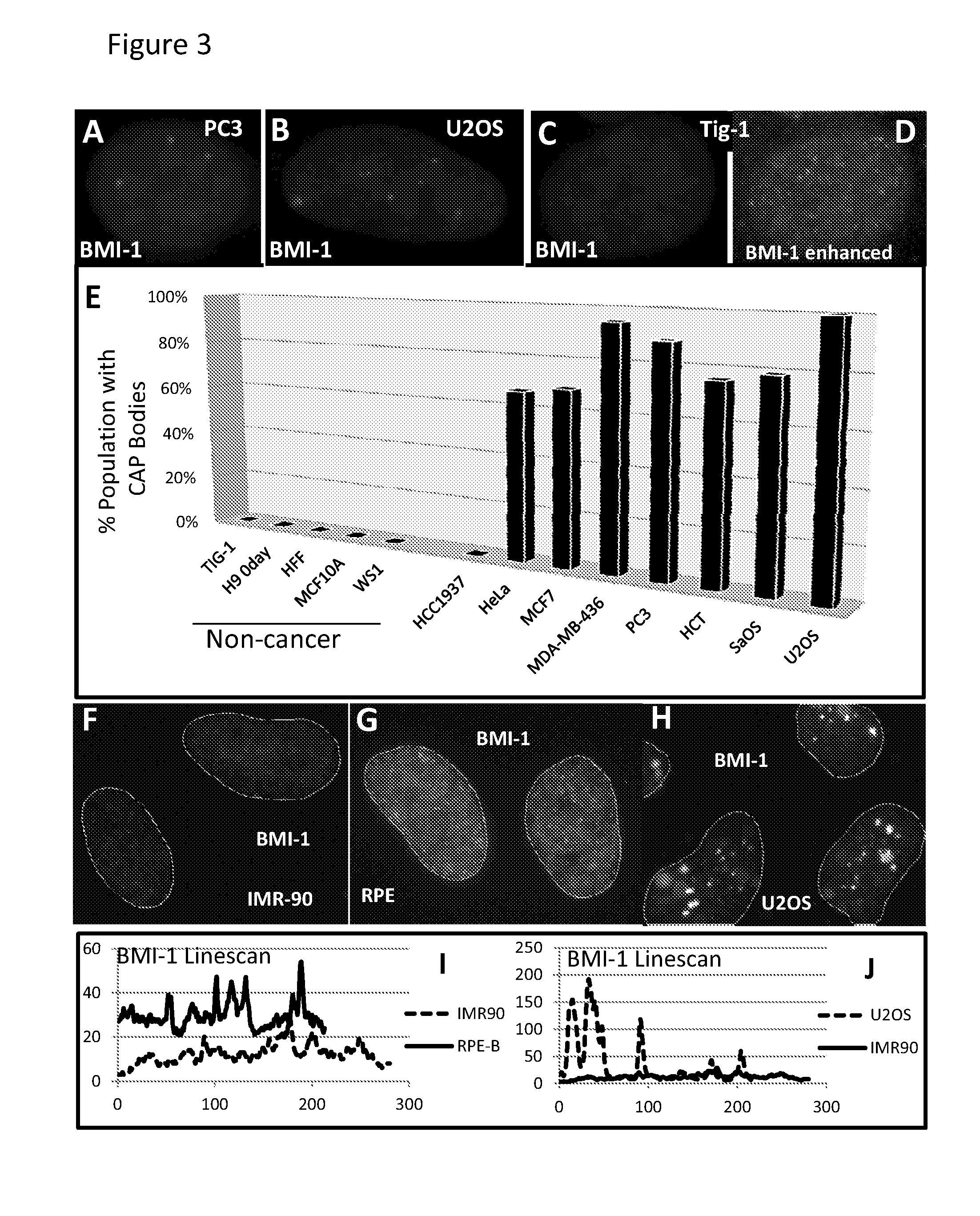
The invention features methods of diagnosing cancer in a mammal (e.g., a human) by detecting a biomarker selected from a satellite II ribonucleic acid (RNA) molecule, a cancer-associated polycomb group (CAP) body, a cancer-associated satellite transcript (CAST) body, and UbH2A. Also featured is a method for identifying an agent for treating cancer in a mammal by contacting a cancer cell having a biomarker selected from a CAP body, a CAST body, and a satellite II RNA molecule with a test agent and determining whether the test agent reduces the level of the biomarker in the cancer cell. Other inventions featured are a method for determining whether a chemotherapeutic agent increases epigenetic imbalance of a cell and a method for detecting epigenetic imbalance by determining a copy number of a satellite II DNA locus at chromosome 1q12 in a cell.
Owner:UNIV OF MASSACHUSETTS
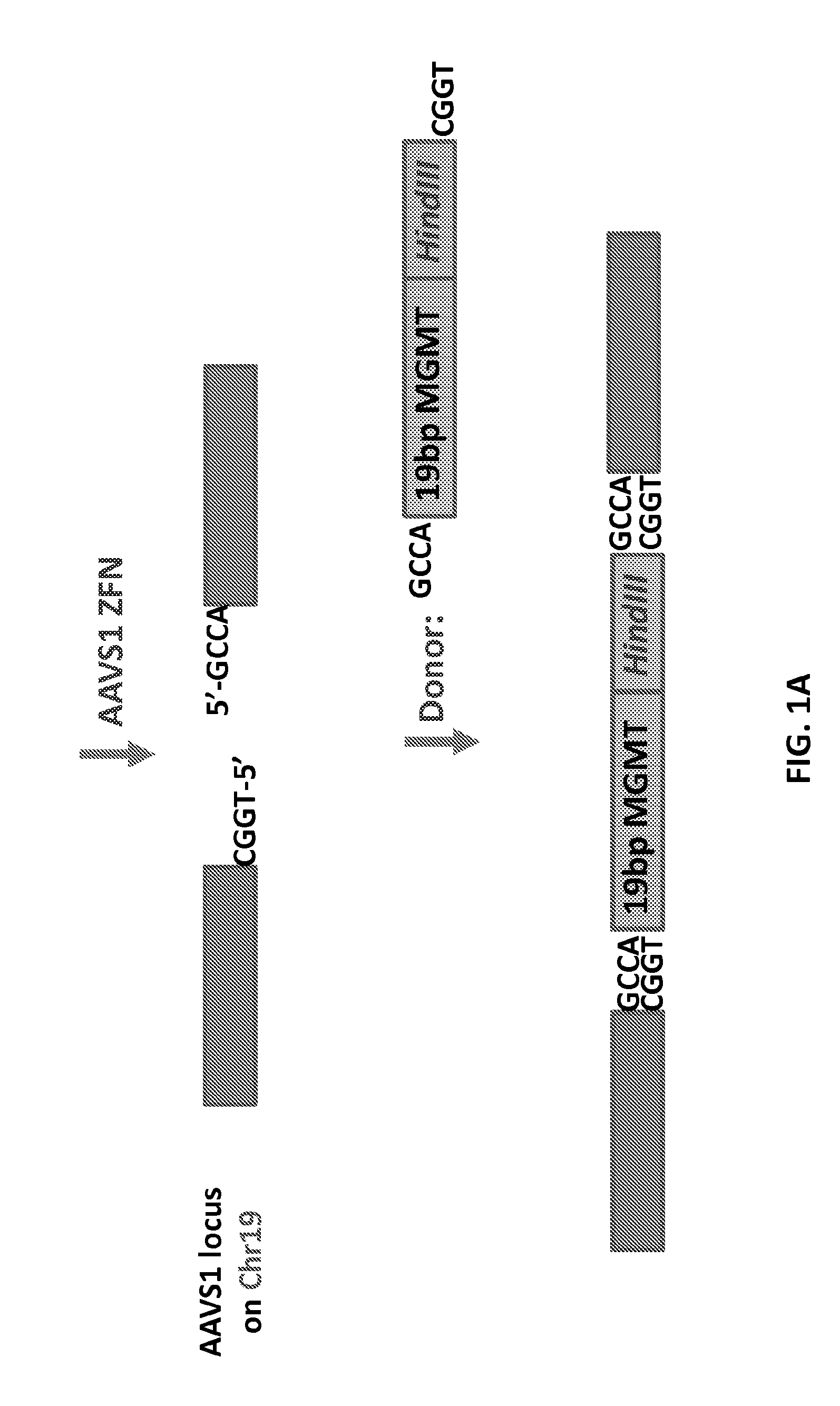
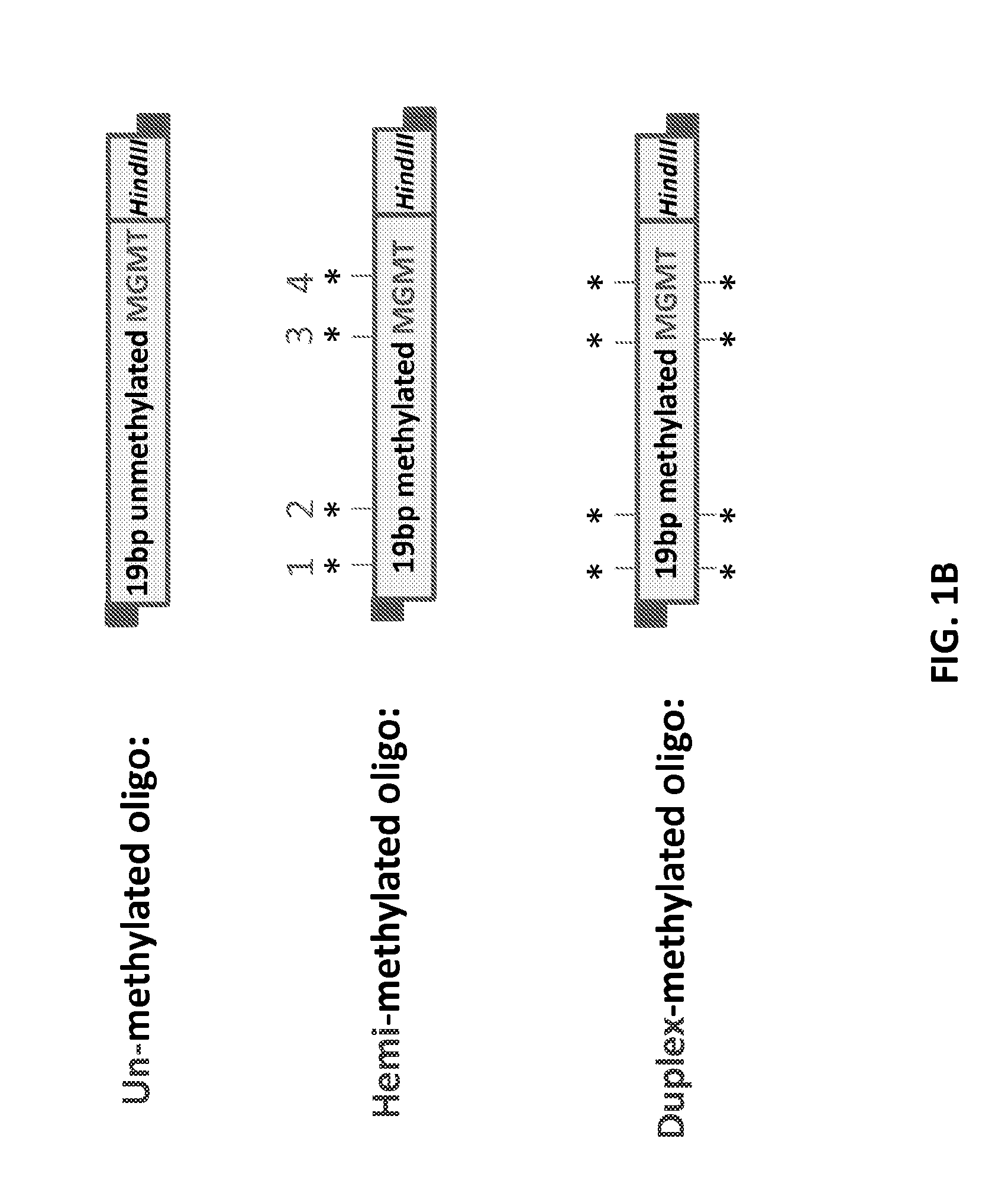
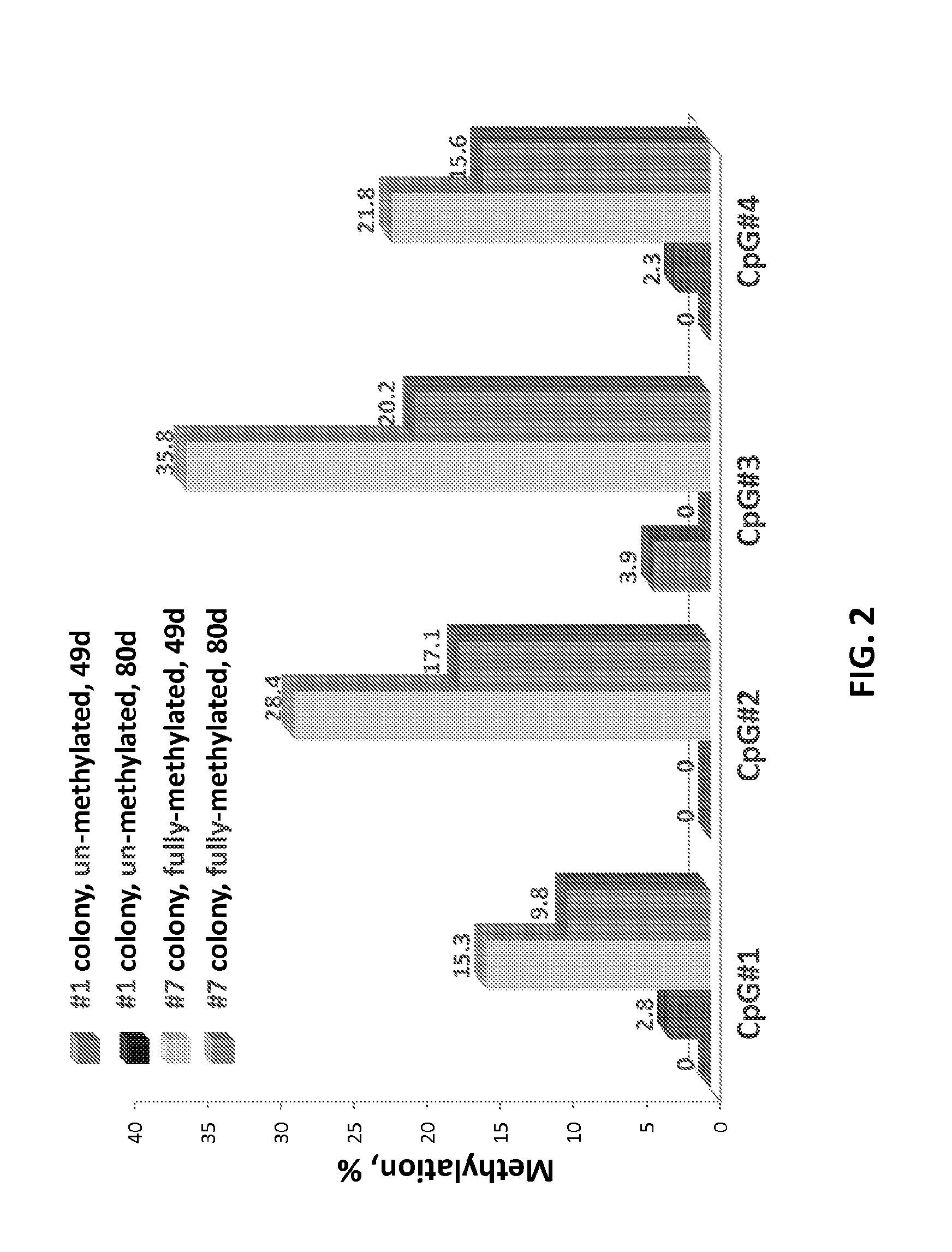
Epigenetic modification of mammalian genomes using targeted endonucleases
The present disclosure provides genetically engineered cell lines comprising chromosomally integrated synthetic sequences having predetermined epigenetic modifications, wherein a predetermined epigenetic modification is correlated with a known diagnosis, prognosis or level of sensitivity to a disease treatment. Also provided are kits comprising said epigenetically modified synthetic nucleic acids or cells comprising said epigenetically modified synthetic nucleic acids that can be used as reference standards for predicting responsiveness to therapeutic treatments, diagnosing diseases, or predicting disease prognosis.
Owner:SIGMA ALDRICH CO LLC
Diagnosis of diseases associated with the cell cycle
InactiveUS20040029123A1Easy to detectBioreactor/fermenter combinationsOrganic active ingredientsCytosineOligomer
The present invention relates to the chemically modified genomic sequences of genes associated with cell cycle, to oligonucleotides and / or PNA-oligomers for detecting the cytosine methylation state of genes associated with cell cycle which are directed against the sequence, as well as to a method for ascertaining genetic and / or epigenetic parameters of genes associated with cell cycle.
Owner:EPIGENOMICS AG
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com