Screening method and application of multiple-allele PCR (Polymerase Chain Reaction) amplified fragment
A technology for amplifying fragments and alleles, applied in the field of molecular biology, can solve the problems of limited information, low cost of industrialization, and few mutations, and achieve high efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
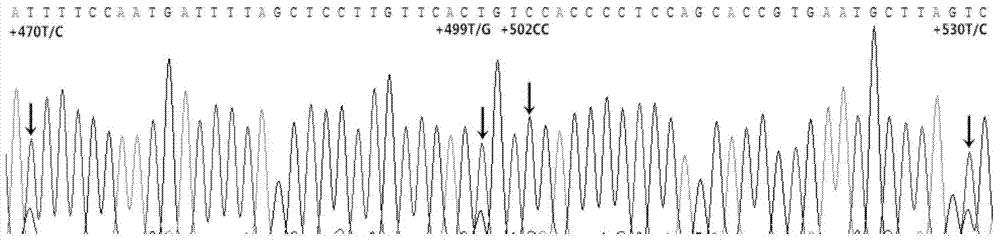
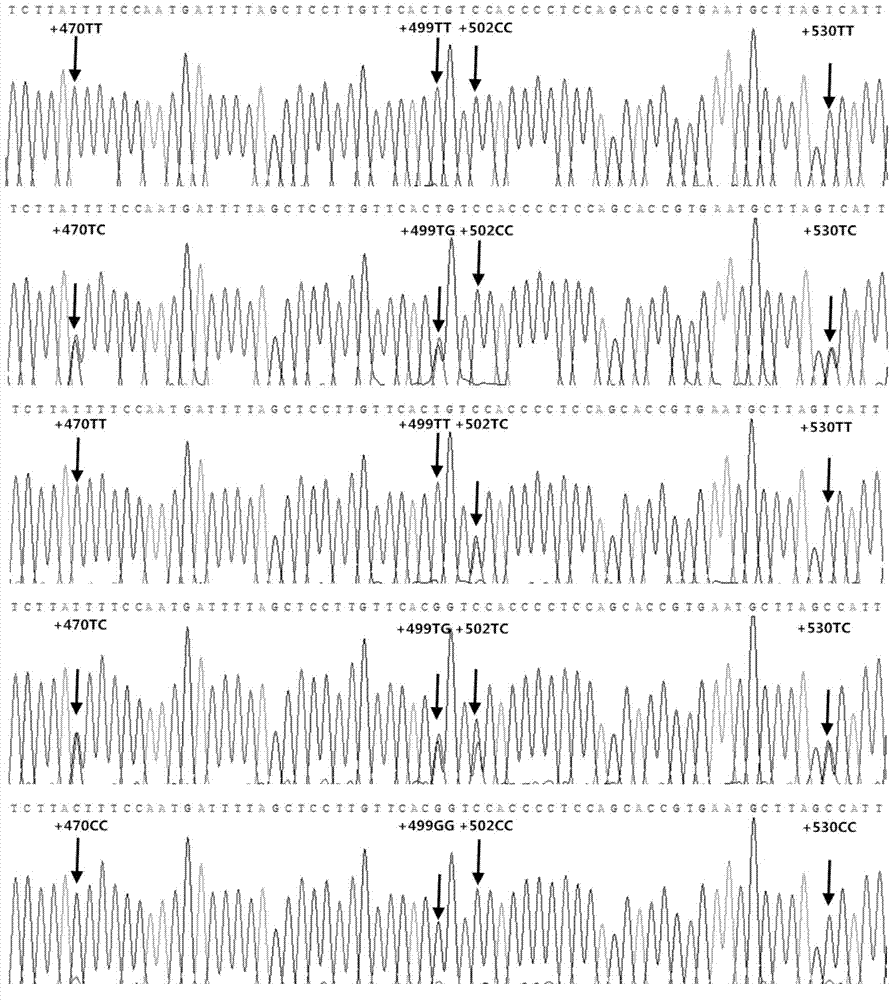
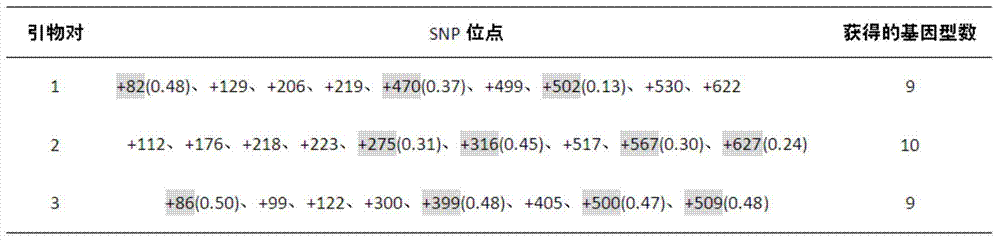
[0017] Example 1 Multi-genotype PCR amplification fragment screening and individual identification of Duroc pig genomic DNA
[0018] 1.1 Primer design (taking pig chromosome 10 as an example)
[0019] 1.1.1 Nucleotide database in NCBI ( http: / / www.ncbi.nlm.nih.gov / nuccore ) in the search keyword Sus scrofa breed mixed chromosome10 (design primers on other chromosomes, change "10" to the corresponding chromosome number). After opening the page, click the hyperlinks Sus scrofa breed mixedchromosome10, Sscrofa10.2, Graphics in turn.
[0020] Click the configure button in the graphical interface to pop up the tab, and among all the options under the Tracks tab, only keep the "√" in front of the sequence and SNP. Click the configure button to confirm.
[0021] 1.1.2 Right-click on the darker blue (SNP dense) place in the SNP barcode, select "Zoom to sequence" from the pop-up menu, and adjust the scale to display in k through the "Zoom bar". Hold down the left mouse button on t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com