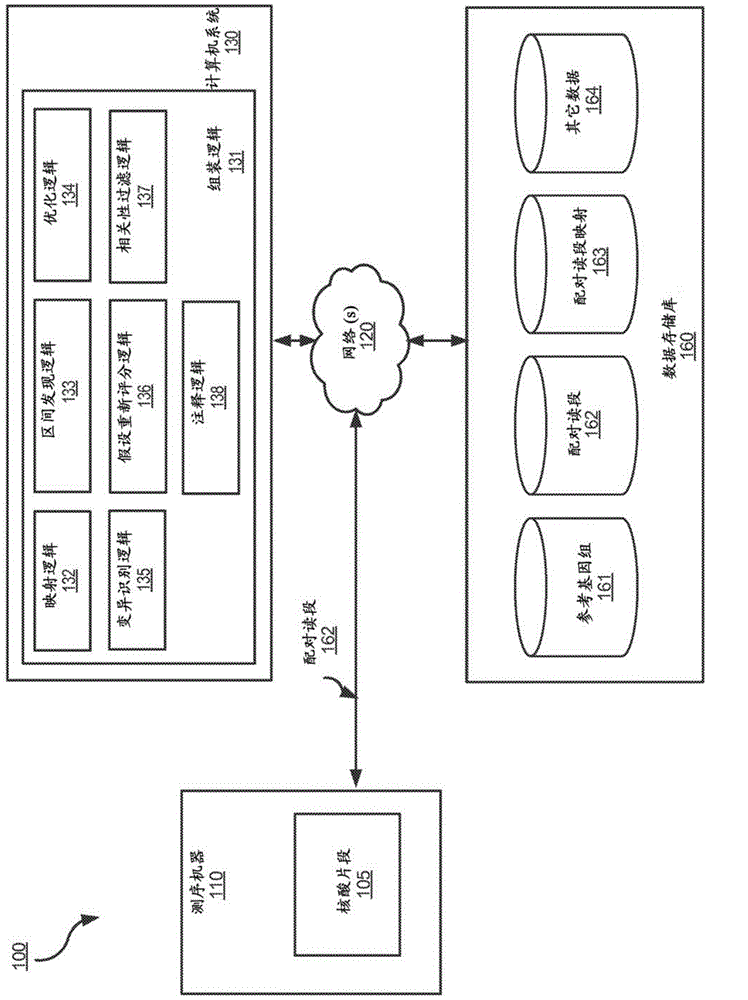
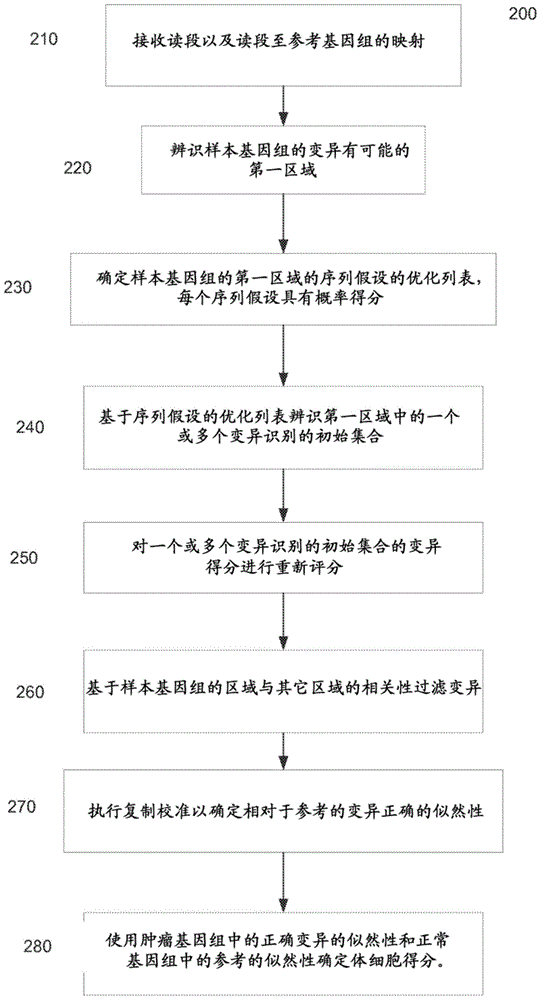
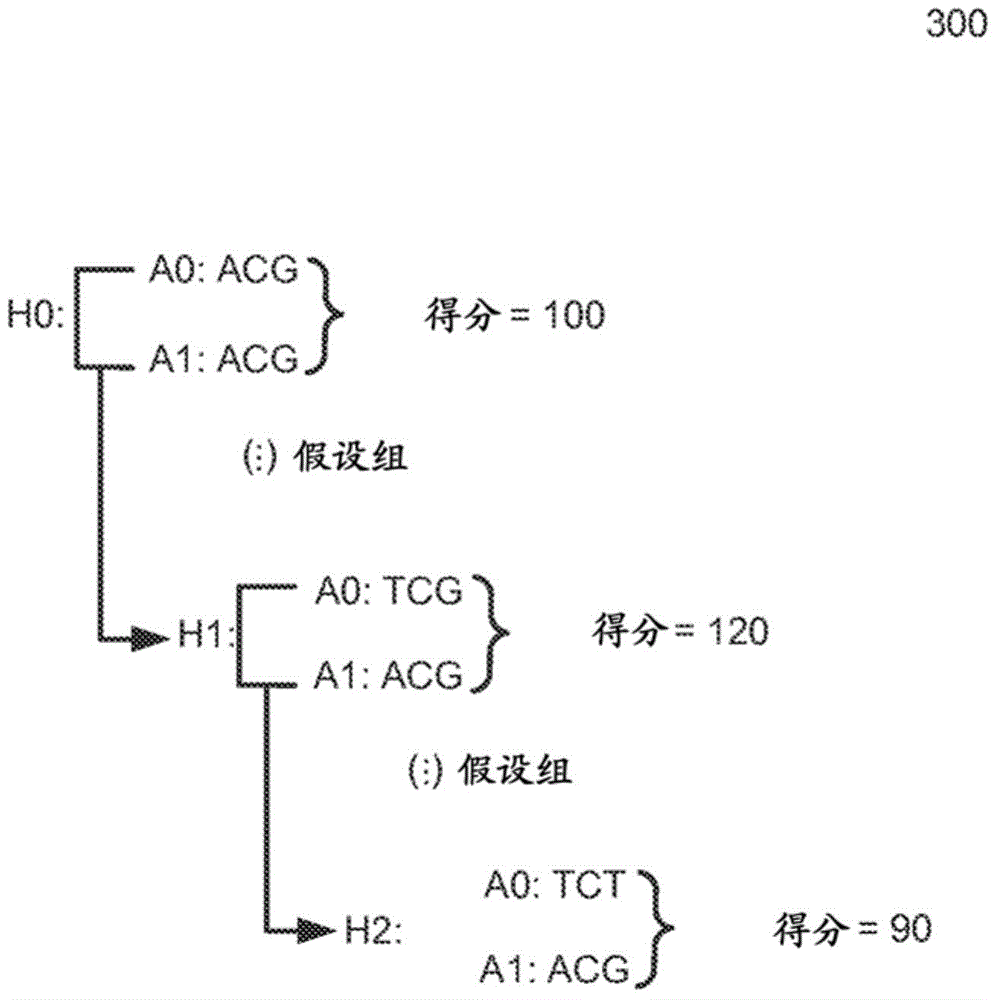
Determining variants in a genome of a heterogeneous sample
A genome and reference genome technology, applied in the field of determining the variation in the genome of heterogeneous samples, can solve all mutation difficulties and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0045] Cancer samples are complex. For example, different cells of a tumor sample may have different genomes. These samples often exhibit this heterogeneity in the genome due to contamination of normal DNA and / or multiple branches in tumor evolution. When these different cells are analyzed within the same sequencing experiment, the measured copy number of the allele at a particular locus may vary. For example, the percentage of DNA with a particular allele (allele fraction) may have any value between 0% and 100%. Therefore, a major challenge in studying cancer genomes is to be able to detect variants present in small fractions of cells in a cancer sample.
[0046] To address this challenge, a process for determining the genome of a sample in a particular region can explicitly allow the allele fraction to vary between ranges of values (eg, any value between 0% and 100%). This determined genome of a sample may effectively be a composite of the genomes of the different cells w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com